Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex
- PMID: 19580872
- PMCID: PMC2757488
- DOI: 10.1016/j.jsb.2009.07.001
Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex
Abstract
The Bacillus subtilis Spx protein is a global transcription factor that interacts with the C-terminal domain of the RNA polymerase alpha subunit (alphaCTD) and regulates transcription of genes involved in thiol-oxidative stress, sporulation, competence, and organosulfur metabolism. Here we determined the X-ray crystal structure of the Spx/alphaCTD complex from an entirely new crystal form than previously reported [Newberry, K.J., Nakano, S., Zuber, P., Brennan, R.G., 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. USA 102, 15839-15844]. Comparison of the previously reported sulfate-bound complex and our sulfate-free complex reveals subtle conformational changes that may be important for the role of Spx in regulating organosulfur metabolism.
Figures
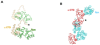
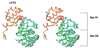

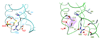
References
-
- Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: The CAP-αCTD-DNA complex. Science. 2002;297:1562–1566. - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. - PubMed
-
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

