In vivo mutational analysis of YtvA from Bacillus subtilis: mechanism of light activation of the general stress response
- PMID: 19581299
- PMCID: PMC2757199
- DOI: 10.1074/jbc.M109.033316
In vivo mutational analysis of YtvA from Bacillus subtilis: mechanism of light activation of the general stress response
Abstract
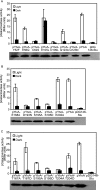
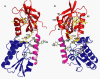
The general stress response of Bacillus subtilis can be activated by stimuli such as the addition of salt or ethanol and with blue light. In the latter response, YtvA activates sigma(B) through a cascade of Rsb proteins, organized in stressosomes. YtvA is composed of an N-terminal LOV (light, oxygen, and voltage) domain and a C-terminal STAS (sulfate transporter and anti-sigma factor) domain and shows light-modulated GTP binding in vitro. Here, we examine the mechanism of YtvA-mediated activation of sigma(B) in vivo with site-directed mutagenesis. Constitutive off and constitutive on mutations have been identified. Disruption of GTP binding in the STAS domain eliminates light activation of sigma(B). In contrast, modification of sites relevant for phosphorylation of STAS domains does not affect the stress response significantly. The data obtained are integrated into a model for the structure of full-length YtvA, which presumably functions as a dimer.
Figures


References
-
- Huala E., Oeller P. W., Liscum E., Han I. S., Larsen E., Briggs W. R. (1997) Science 278, 2120–2123 - PubMed
-
- Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., Briggs W. R. (1998) Science 282, 1698–1701 - PubMed
-
- Losi A. (2007) Photochem. Photobiol. 83, 1283–1300 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

