Curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin
- PMID: 19581445
- PMCID: PMC2746146
- DOI: 10.1534/genetics.109.105601
Curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin
Abstract
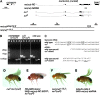
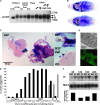
Drosophila melanogaster curled, one of the first fly mutants described by T. H. Morgan >90 years ago, is the founding member of a series of curled wing phenotype mutants widely used as markers in fruit fly genetics. The expressivity of the wing phenotype is environmentally modulated, suggesting that the mutation affects the metabolic status of cells rather than a developmental control gene. However, the molecular identity of any of the curled wing marker mutant genes is still unknown. In a screen for starvation-responsive genes, we previously identified the single fly homolog of the vertebrate nocturnin genes, which encode cytoplasmic deadenylases that act in the post-transcriptional control of genes by poly(A) tail removal of target mRNAs prior to their degradation. Here we show that curled encodes Drosophila Nocturnin and that the gene is required at pupal stage for proper wing morphogenesis after eclosion of the fly. Despite the complex ontogenetic expression pattern of the gene, curled is not expressed in the developing wing, and wing-specific curled knockdown mediated by RNAi does not result in the curled wing phenotype, indicating a tissue-nonautonomous, systemic mode of curled gene function. Our study not only presents an entry point into the functional analysis of invertebrate nocturnins but also paves the way for the identification of the still elusive Nocturnin target mRNAs by genetic suppressor screens on the curled wing phenotype.
Figures

References
-
- Baggs, J. E., and C. B. Green, 2003. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13: 189–198. - PubMed
-
- Baker, J. D., and J. W. Truman, 2002. Mutations in the Drosophila glycoprotein hormone receptor, rickets, eliminate neuropeptide-induced tanning and selectively block a stereotyped behavioral program. J. Exp. Biol. 205: 2555–2565. - PubMed
-
- Ball, F., 1935. Mutants and abberations. Drosoph. Inf. Serv. 3: 108.
-
- Barbot, W., M. Wasowicz, A. Dupressoir, C. Versaux-Botteri and T. Heidmann, 2002. A murine gene with circadian expression revealed by transposon insertion: self-sustained rhythmicity in the liver and the photoreceptors. Biochim. Biophys. Acta 1576: 81–91. - PubMed
-
- Bönisch, C., C. Temme, B. Moritz and E. Wahle, 2007. Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J. Biol. Chem. 282: 21818–21828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

