Role of dimerization in the catalytic properties of the Escherichia coli disulfide isomerase DsbC
- PMID: 19581640
- PMCID: PMC2781991
- DOI: 10.1074/jbc.M109.010199
Role of dimerization in the catalytic properties of the Escherichia coli disulfide isomerase DsbC
Abstract
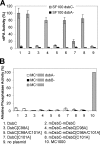
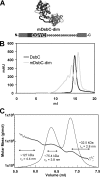
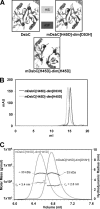
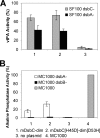
The bacterial protein-disulfide isomerase DsbC is a homodimeric V-shaped enzyme that consists of a dimerization domain, two alpha-helical linkers, and two opposing thioredoxin fold catalytic domains. The functional significance of the two catalytic domains of DsbC is not well understood yet. We have engineered heterodimer-like DsbC derivatives covalently linked via (Gly(3)-Ser) flexible linkers. We either inactivated one of the catalytic sites (CGYC), or entirely removed one of the catalytic domains while maintaining the putative binding area intact. Variants having a single active catalytic site display significant levels of isomerase activity. Furthermore, mDsbC[H45D]-dim[D53H], a DsbC variant lacking an entire catalytic domain but with an intact dimerization domain, also showed isomerase activity, albeit at lower levels. In addition, the absence of the catalytic domain allowed this protein to catalyze in vivo oxidation. Our results reveal that two catalytic domains in DsbC are not essential for disulfide bond isomerization and that a determining feature in isomerization is the availability of a substrate binding domain.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

