Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells
- PMID: 19582141
- PMCID: PMC2700273
- DOI: 10.1371/journal.pbio.1000149
Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells
Abstract
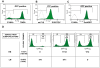
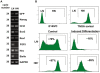
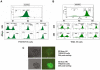
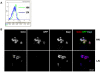
There is evidence that pluripotency of mouse embryonic stem (ES) cells is associated with the activity of a network of transcription factors with Sox2, Oct4, and Nanog at the core. Using fluorescent reporters for the expression of Nanog, we observed that a population of ES cells is best described by a dynamic distribution of Nanog expression characterized by two peaks defined by high (HN) and low (LN) Nanog expression. Typically, the LN state is 5%-20% of the total population, depending on the culture conditions. Modelling of the activity of Nanog reveals that a simple network of Oct4/Sox2 and Nanog activity can account for the observed distribution and its properties as long as the transcriptional activity is tuned by transcriptional noise. The model also predicts that the LN state is unstable, something that is born out experimentally. While in this state, cells can differentiate. We suggest that transcriptional fluctuations in Nanog expression are an essential element of the pluripotent state and that the function of Sox2, Oct4, and Nanog is to act as a network that promotes and maintains transcriptional noise to interfere with the differentiation signals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
-
- Smith AG. Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. - PubMed
-
- Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

