Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a
- PMID: 19582148
- PMCID: PMC2701631
- DOI: 10.1371/journal.pone.0006161
Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a
Abstract
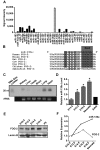
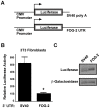
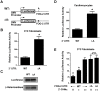
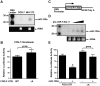
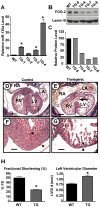
MicroRNAs are increasingly being recognized as regulators of embryonic development; however, relatively few microRNAs have been identified to regulate cardiac development. FOG-2 (also known as zfpm2) is a transcriptional co-factor that we have previously shown is critical for cardiac development. In this report, we demonstrate that FOG-2 expression is controlled at the translational level by microRNA-130a. We identified a conserved region in the FOG-2 3' untranslated region predicted to be a target for miR-130a. To test the functional significance of this site, we generated an expression construct containing the luciferase coding region fused with the 3' untranslated region of FOG-2 or a mutant version lacking this microRNA binding site. When these constructs were transfected into NIH 3T3 fibroblasts (which are known to express miR-130a), we observed a 3.3-fold increase in translational efficiency when the microRNA target site was disrupted. Moreover, knockdown of miR-130a in fibroblasts resulted in a 3.6-fold increase in translational efficiency. We also demonstrate that cardiomyocytes express miR-130a and can attenuate translation of mRNAs with a FOG-2 3' untranslated region. Finally, we generated transgenic mice with cardiomyocyte over-expression of miR-130a. In the hearts of these mice, FOG-2 protein levels were reduced by as much as 80%. Histological analysis of transgenic embryos revealed ventricular wall hypoplasia and ventricular septal defects, similar to that seen in FOG-2 deficient hearts. These results demonstrate the importance of miR-130a for the regulation of FOG-2 protein expression and suggest that miR-130a may also play a role in the regulation of cardiac development.
Conflict of interest statement
Figures
Similar articles
-
The miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation of mouse embryonic cardiomyocytes.Braz J Med Biol Res. 2012 Feb;45(2):131-8. doi: 10.1590/s0100-879x2012007500007. Epub 2012 Feb 9. Braz J Med Biol Res. 2012. PMID: 22267003 Free PMC article.
-
Downregulation of connexin43 by microRNA-130a in cardiomyocytes results in cardiac arrhythmias.J Mol Cell Cardiol. 2014 Sep;74:53-63. doi: 10.1016/j.yjmcc.2014.04.024. Epub 2014 May 10. J Mol Cell Cardiol. 2014. PMID: 24819345 Free PMC article.
-
MicroRNA-130a Regulation of Desmocollin 2 in a Novel Model of Arrhythmogenic Cardiomyopathy.Microrna. 2017;6(2):143-150. doi: 10.2174/2211536605666161109111031. Microrna. 2017. PMID: 27834139 Free PMC article.
-
MicroRNA-130a is up-regulated in mouse liver by iron deficiency and targets the bone morphogenetic protein (BMP) receptor ALK2 to attenuate BMP signaling and hepcidin transcription.J Biol Chem. 2014 Aug 22;289(34):23796-808. doi: 10.1074/jbc.M114.577387. Epub 2014 Jul 7. J Biol Chem. 2014. PMID: 25002578 Free PMC article.
-
MicroRNA-130a, a Potential Antifibrotic Target in Cardiac Fibrosis.J Am Heart Assoc. 2017 Nov 7;6(11):e006763. doi: 10.1161/JAHA.117.006763. J Am Heart Assoc. 2017. PMID: 29114000 Free PMC article.
Cited by
-
MicroRNAs: new players in heart failure.Mol Biol Rep. 2013 Mar;40(3):2663-70. doi: 10.1007/s11033-012-2352-y. Epub 2012 Dec 15. Mol Biol Rep. 2013. PMID: 23242657 Review.
-
Increased HDAC3 and decreased miRNA-130a expression in PBMCs through recruitment HDAC3 in patients with spinal cord injuries.Int J Clin Exp Pathol. 2015 Feb 1;8(2):1682-9. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 25973054 Free PMC article.
-
Integrated small RNA, mRNA and protein omics reveal a miRNA network orchestrating metabolic maturation of the developing human heart.BMC Genomics. 2023 Nov 23;24(1):709. doi: 10.1186/s12864-023-09801-8. BMC Genomics. 2023. PMID: 37996818 Free PMC article.
-
MicroRNA-133a engineered mesenchymal stem cells augment cardiac function and cell survival in the infarct heart.J Cardiovasc Pharmacol. 2015 Mar;65(3):241-51. doi: 10.1097/FJC.0000000000000183. J Cardiovasc Pharmacol. 2015. PMID: 25658461 Free PMC article.
-
MicroRNA-130a Increases and Predicts Cardiotoxicity during Adjuvant Chemotherapy in Human Epidermal Growth Factor Receptor-2-Positive Breast Cancer.J Breast Cancer. 2021 Apr;24(2):153-163. doi: 10.4048/jbc.2021.24.e15. Epub 2021 Mar 2. J Breast Cancer. 2021. PMID: 33818020 Free PMC article.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. - PubMed
-
- Sontheimer EJ, Carthew RW. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. - PubMed
-
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. - PubMed
-
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

