Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery
- PMID: 19582160
- PMCID: PMC2702084
- DOI: 10.1371/journal.pone.0006146
Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery
Abstract
Background: Breast cancer cell lines have been used widely to investigate breast cancer pathobiology and new therapies. Breast cancer is a molecularly heterogeneous disease, and it is important to understand how well and which cell lines best model that diversity. In particular, microarray studies have identified molecular subtypes-luminal A, luminal B, ERBB2-associated, basal-like and normal-like-with characteristic gene-expression patterns and underlying DNA copy number alterations (CNAs). Here, we studied a collection of breast cancer cell lines to catalog molecular profiles and to assess their relation to breast cancer subtypes.
Methods: Whole-genome DNA microarrays were used to profile gene expression and CNAs in a collection of 52 widely-used breast cancer cell lines, and comparisons were made to existing profiles of primary breast tumors. Hierarchical clustering was used to identify gene-expression subtypes, and Gene Set Enrichment Analysis (GSEA) to discover biological features of those subtypes. Genomic and transcriptional profiles were integrated to discover within high-amplitude CNAs candidate cancer genes with coordinately altered gene copy number and expression.
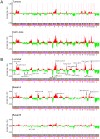
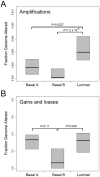
Findings: Transcriptional profiling of breast cancer cell lines identified one luminal and two basal-like (A and B) subtypes. Luminal lines displayed an estrogen receptor (ER) signature and resembled luminal-A/B tumors, basal-A lines were associated with ETS-pathway and BRCA1 signatures and resembled basal-like tumors, and basal-B lines displayed mesenchymal and stem/progenitor-cell characteristics. Compared to tumors, cell lines exhibited similar patterns of CNA, but an overall higher complexity of CNA (genetically simple luminal-A tumors were not represented), and only partial conservation of subtype-specific CNAs. We identified 80 high-level DNA amplifications and 13 multi-copy deletions, and the resident genes with concomitantly altered gene-expression, highlighting known and novel candidate breast cancer genes.
Conclusions: Overall, breast cancer cell lines were genetically more complex than tumors, but retained expression patterns with relevance to the luminal-basal subtype distinction. The compendium of molecular profiles defines cell lines suitable for investigations of subtype-specific pathobiology, cancer stem cell biology, biomarkers and therapies, and provides a resource for discovery of new breast cancer genes.
Conflict of interest statement
Figures


References
-
- Subramaniam DS, Isaacs C. Utilizing prognostic and predictive factors in breast cancer. Curr Treat Options Oncol. 2005;6:147–159. - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. - PubMed
-
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

