Towards inferring time dimensionality in protein-protein interaction networks by integrating structures: the p53 example
- PMID: 19585003
- PMCID: PMC2898629
- DOI: 10.1039/B905661K
Towards inferring time dimensionality in protein-protein interaction networks by integrating structures: the p53 example
Abstract
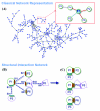
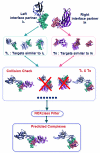
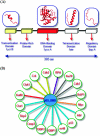
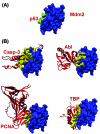
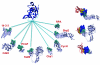
Inspection of protein-protein interaction maps illustrates that a hub protein can interact with a very large number of proteins, reaching tens and even hundreds. Since a single protein cannot interact with such a large number of partners at the same time, this presents a challenge: can we figure out which interactions can occur simultaneously and which are mutually excluded? Addressing this question adds a fourth dimension into interaction maps: that of time. Including the time dimension in structural networks is an immense asset; time dimensionality transforms network node-and-edge maps into cellular processes, assisting in the comprehension of cellular pathways and their regulation. While the time dimensionality can be further enhanced by linking protein complexes to time series of mRNA expression data, current robust, network experimental data are lacking. Here we outline how, using structural data, efficient structural comparison algorithms and appropriate datasets and filters can assist in getting an insight into time dimensionality in interaction networks; in predicting which interactions can and cannot co-exist; and in obtaining concrete predictions consistent with experiment. As an example, we present p53-linked processes.
Figures
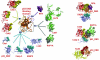


Similar articles
-
P53 mdm2 inhibitors.Curr Pharm Des. 2012;18(30):4668-78. doi: 10.2174/138161212802651580. Curr Pharm Des. 2012. PMID: 22650254 Free PMC article. Review.
-
Functional profiling of p53-binding sites in Hdm2 and Hdmx using a genetic selection system.Bioorg Med Chem. 2010 Aug 15;18(16):6099-108. doi: 10.1016/j.bmc.2010.06.053. Epub 2010 Jun 22. Bioorg Med Chem. 2010. PMID: 20638853 Free PMC article.
-
Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway.PLoS Biol. 2006 Feb;4(2):e27. doi: 10.1371/journal.pbio.0040027. Epub 2006 Jan 17. PLoS Biol. 2006. PMID: 16402859 Free PMC article.
-
iSEE: Interface structure, evolution, and energy-based machine learning predictor of binding affinity changes upon mutations.Proteins. 2019 Feb;87(2):110-119. doi: 10.1002/prot.25630. Epub 2018 Dec 3. Proteins. 2019. PMID: 30417935 Free PMC article.
-
Small molecule inhibitors of the p53-MDM2.Curr Med Chem. 2008;15(17):1720-30. doi: 10.2174/092986708784872375. Curr Med Chem. 2008. PMID: 18673221 Review.
Cited by
-
QSLiMFinder: improved short linear motif prediction using specific query protein data.Bioinformatics. 2015 Jul 15;31(14):2284-93. doi: 10.1093/bioinformatics/btv155. Epub 2015 Mar 19. Bioinformatics. 2015. PMID: 25792551 Free PMC article.
-
Novel insights through the integration of structural and functional genomics data with protein networks.J Struct Biol. 2012 Sep;179(3):320-6. doi: 10.1016/j.jsb.2012.02.001. Epub 2012 Feb 11. J Struct Biol. 2012. PMID: 22343087 Free PMC article.
-
Impact of Alu repeats on the evolution of human p53 binding sites.Biol Direct. 2011 Jan 6;6:2. doi: 10.1186/1745-6150-6-2. Biol Direct. 2011. PMID: 21208455 Free PMC article.
-
Cryptochrome deletion in p53 mutant mice enhances apoptotic and anti-tumorigenic responses to UV damage at the transcriptome level.Funct Integr Genomics. 2019 Sep;19(5):729-742. doi: 10.1007/s10142-019-00680-5. Epub 2019 May 1. Funct Integr Genomics. 2019. PMID: 31044344
-
Roles of computational modelling in understanding p53 structure, biology, and its therapeutic targeting.J Mol Cell Biol. 2019 Apr 1;11(4):306-316. doi: 10.1093/jmcb/mjz009. J Mol Cell Biol. 2019. PMID: 30726928 Free PMC article. Review.
References
-
- Aloy P., Pichaud M., Russell R. B. Curr. Opin. Struct. Biol. 2005;15(1):15–22. - PubMed
-
- Aloy P., Russell R. B. Trends Biochem. Sci. 2002;27(12):633–638. - PubMed
-
- Aloy P., Russell R. B. Nat. Rev. Mol. Cell Biol. 2006;7(3):188–197. - PubMed
-
- Bork P., Serrano L. Cell (Cambridge, Mass.) 2005;121(4):507–509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

