Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A
- PMID: 19586908
- PMCID: PMC2782015
- DOI: 10.1074/jbc.M109.000745
Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A
Abstract
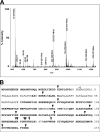
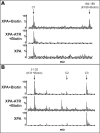
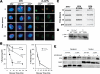
In response to DNA damage, eukaryotic cells activate a series of DNA damage-dependent pathways that serve to arrest cell cycle progression and remove DNA damage. Coordination of cell cycle arrest and damage repair is critical for maintenance of genomic stability. However, this process is still poorly understood. Nucleotide excision repair (NER) and the ATR-dependent cell cycle checkpoint are the major pathways responsible for repair of UV-induced DNA damage. Here we show that ATR physically interacts with the NER factor Xeroderma pigmentosum group A (XPA). Using a mass spectrometry-based protein footprinting method, we found that ATR interacts with a helix-turn-helix motif in the minimal DNA-binding domain of XPA where an ATR phosphorylation site (serine 196) is located. XPA-deficient cells complemented with XPA containing a point mutation of S196A displayed a reduced repair efficiency of cyclobutane pyrimidine dimers as compared with cells complemented with wild-type XPA, although no effect was observed for repair of (6-4) photoproducts. This suggests that the ATR-dependent phosphorylation of XPA may promote NER repair of persistent DNA damage. In addition, a K188A point mutation of XPA that disrupts the ATR-XPA interaction inhibits the nuclear import of XPA after UV irradiation and, thus, significantly reduced DNA repair efficiency. By contrast, the S196A mutation has no effect on XPA nuclear translocation. Taken together, our results suggest that the ATR-XPA interaction mediated by the helix-turn-helix motif of XPA plays an important role in DNA-damage responses to promote cell survival and genomic stability after UV irradiation.
Figures



References
-
- Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 - PubMed
-
- Park C. J., Choi B. S. (2006) FEBS J. 273, 1600–1608 - PubMed
-
- Kraemer K. H., Lee M. M., Andrews A. D., Lambert W. C. (1994) Arch. Dermatol. 130, 1018–1021 - PubMed
-
- Kraemer K. H., Lee M. M., Scotto J. (1987) Arch. Dermatol. 123, 241–250 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

