Inactive X chromosome-specific reduction in placental DNA methylation
- PMID: 19586922
- PMCID: PMC2742397
- DOI: 10.1093/hmg/ddp299
Inactive X chromosome-specific reduction in placental DNA methylation
Abstract
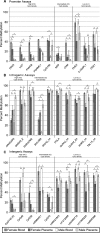
Genome-wide levels of DNA methylation vary between tissues, and compared with other tissues, the placenta has been reported to demonstrate a global decrease in methylation as well as decreased methylation of X-linked promoters. Methylation is one of many features that differentiate the active and inactive X, and it is well established that CpG island promoters on the inactive X are hypermethylated. We now report a detailed analysis of methylation at different regions across the X in male and female placenta and blood. A significant (P < 0.001) placental hypomethylation of LINE1 elements was observed in both males and females. Relative to blood placental promoter hypomethylation was only observed for X-linked, not autosomal promoters, and was significant for females (P < 0.0001) not males (P = 0.9266). In blood, X-linked CpG island promoters were shown to have moderate female methylation (66% across 70 assays) and low (23%) methylation in males. A similar methylation pattern in blood was observed for approximately 20% of non-island promoters as well as 50% of the intergenic or intragenic CpG islands, the latter is likely due to the presence of unannotated promoters. Both intragenic and intergenic regions showed similarly high methylation levels in male and female blood (68 and 66%) while placental methylation of these regions was lower, particularly in females. Thus placental hypomethylation relative to blood is observed globally at repetitive elements as well as across the X. The decrease in X-linked placental methylation is consistently greater in females than males and implicates an inactive X specific loss of methylation in the placenta.
Figures



References
-
- Gama-Sosa M.A., Midgett R.M., Slagel V.A., Githens S., Kuo K.C., Gehrke C.W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta. 1983;740:212–219. - PubMed
-
- Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. - PubMed
-
- Shen H.M., Nakamura A., Sugimoto J., Sakumoto N., Oda T., Jinno Y., Okazaki Y. Tissue specificity of methylation and expression of human genes coding for neuropeptides and their receptors, and of a human endogenous retrovirus K family. J. Hum. Genet. 2006;51:440–450. - PubMed

