Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants
- PMID: 19587035
- PMCID: PMC2738245
- DOI: 10.1128/JVI.02076-08
Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants
Abstract
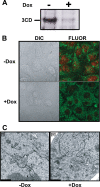
A poliovirus (PV) mutant (termed GG), which is incapable of producing 3AB, VPg, and 3CD proteins due to a defective cleavage site between the 3B and 3C proteins, replicated, producing 3BC-linked RNA rather than the VPg-linked RNA produced by the wild type (WT). GG PV RNA is quasi-infectious. The yield of infectious GG PV relative to replicated RNA is reduced by almost 5 logs relative to that of WT PV. Proteolytic activity required for polyprotein processing is normal for the GG mutant. 3BC-linked RNA can be encapsidated as efficiently as VPg-linked RNA. However, a step after genome replication but preceding virus assembly that is dependent on 3CD and/or 3AB proteins limits production of infectious GG PV. This step may involve release of replicated genomes from replication complexes. A pseudorevertant (termed EG) partially restored cleavage at the 3B-3C cleavage site. The reduced rate of formation of 3AB and 3CD caused corresponding reductions in the observed rate of genome replication and infectious virus production by EG PV without impacting the final yield of replicated RNA or infectious virus relative to that of WT PV. Using EG PV, we showed that genome replication and encapsidation were distinct steps in the multiplication cycle. Ectopic expression of 3CD protein reversed the genome replication phenotype without alleviating the infectious-virus production phenotype. This is the first report of a trans-complementable function for 3CD for any picornavirus. This observation supports an interaction between 3CD protein and viral and/or host factors that is critical for genome replication, perhaps formation of replication complexes.
Figures


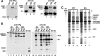


Similar articles
-
An efficient trans complementation system for in vivo replication of defective poliovirus mutants.J Virol. 2024 Jul 23;98(7):e0052324. doi: 10.1128/jvi.00523-24. Epub 2024 Jun 5. J Virol. 2024. PMID: 38837378 Free PMC article.
-
Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein, 3AB.J Virol. 2007 Jun;81(11):5669-84. doi: 10.1128/JVI.02350-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360746 Free PMC article.
-
Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex.J Biol Chem. 2008 Jan 11;283(2):875-88. doi: 10.1074/jbc.M707907200. Epub 2007 Nov 9. J Biol Chem. 2008. PMID: 17993457 Free PMC article.
-
IRES-controlled protein synthesis and genome replication of poliovirus.Arch Virol Suppl. 1994;9:279-89. doi: 10.1007/978-3-7091-9326-6_28. Arch Virol Suppl. 1994. PMID: 8032259 Review.
-
Expanding knowledge of P3 proteins in the poliovirus lifecycle.Future Microbiol. 2010 Jun;5(6):867-81. doi: 10.2217/fmb.10.40. Future Microbiol. 2010. PMID: 20521933 Free PMC article. Review.
Cited by
-
Multiple poliovirus-induced organelles suggested by comparison of spatiotemporal dynamics of membranous structures and phosphoinositides.PLoS Pathog. 2018 Apr 27;14(4):e1007036. doi: 10.1371/journal.ppat.1007036. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29702686 Free PMC article.
-
Protein Nucleotidylylation in +ssRNA Viruses.Viruses. 2021 Aug 5;13(8):1549. doi: 10.3390/v13081549. Viruses. 2021. PMID: 34452414 Free PMC article. Review.
-
Viral protein engagement of GBF1 induces host cell vulnerability through synthetic lethality.J Cell Biol. 2022 Nov 7;221(11):e202011050. doi: 10.1083/jcb.202011050. Epub 2022 Oct 28. J Cell Biol. 2022. PMID: 36305789 Free PMC article.
-
Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis.PLoS Pathog. 2010 Aug 26;6(8):e1001066. doi: 10.1371/journal.ppat.1001066. PLoS Pathog. 2010. PMID: 20865167 Free PMC article.
-
Hijacking of multiple phospholipid biosynthetic pathways and induction of membrane biogenesis by a picornaviral 3CD protein.PLoS Pathog. 2018 May 21;14(5):e1007086. doi: 10.1371/journal.ppat.1007086. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29782554 Free PMC article.
References
-
- Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. - PubMed
-
- Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. - PubMed
-
- Amineva, S. P., A. G. Aminev, A. C. Palmenberg, and J. E. Gern. 2004. Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J. Gen. Virol. 85:2969-2979. - PubMed
-
- Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

