Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets
- PMID: 19587057
- PMCID: PMC2738220
- DOI: 10.1128/JVI.01089-09
Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets
Abstract
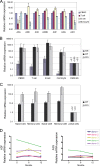
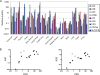
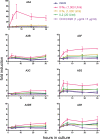
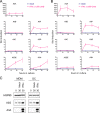
Human APOBEC3 enzymes are cellular DNA cytidine deaminases that inhibit and/or mutate a variety of retroviruses, retrotransposons, and DNA viruses. Here, we report a detailed examination of human APOBEC3 gene expression, focusing on APOBEC3G (A3G) and APOBEC3F (A3F), which are potent inhibitors of human immunodeficiency virus type 1 (HIV-1) infection but are suppressed by HIV-1 Vif. A3G and A3F are expressed widely in hematopoietic cell populations, including T cells, B cells, and myeloid cells, as well as in tissues where mRNA levels broadly correlate with the lymphoid cell content (gonadal tissues are exceptions). By measuring mRNA copy numbers, we find that A3G mRNA is approximately 10-fold more abundant than A3F mRNA, implying that A3G is the more significant anti-HIV-1 factor in vivo. A3G and A3F levels also vary between donors, and these differences are sustained over 12 months. Responses to T-cell activation or cytokines reveal that A3G and A3F mRNA levels are induced approximately 10-fold in macrophages and dendritic cells (DCs) by alpha interferon (IFN-alpha) and approximately 4-fold in naïve CD4(+) T cells. However, immunoblotting revealed that A3G protein levels are induced by IFN-alpha in macrophages and DCs but not in T cells. In contrast, T-cell activation and IFN-gamma had a minimal impact on A3G or A3F expression. Finally, we noted that A3A mRNA expression and protein expression are exquisitely sensitive to IFN-alpha induction in CD4(+) T cells, macrophages, and DCs but not to T-cell activation or other cytokines. Given that A3A does not affect HIV-1 infection, these observations imply that this protein may participate in early antiviral innate immune responses.
Figures


References
-
- An, P., G. Bleiber, P. Duggal, G. Nelson, M. May, B. Mangeat, I. Alobwede, D. Trono, D. Vlahov, S. Donfield, J. J. Goedert, J. Phair, S. Buchbinder, S. J. O'Brien, A. Telenti, and C. A. Winkler. 2004. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 78:11070-11076. - PMC - PubMed
-
- Biasin, M., L. Piacentini, S. Lo Caputo, Y. Kanari, G. Magri, D. Trabattoni, V. Naddeo, L. Lopalco, A. Clivio, E. Cesana, F. Fasano, C. Bergamaschi, F. Mazzotta, M. Miyazawa, and M. Clerici. 2007. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J. Infect. Dis. 195:960-964. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

