Structural and enzymatic characterization of Os3BGlu6, a rice beta-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides
- PMID: 19587102
- PMCID: PMC2735989
- DOI: 10.1104/pp.109.139436
Structural and enzymatic characterization of Os3BGlu6, a rice beta-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides
Abstract
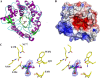
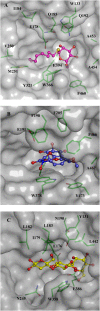
Glycoside hydrolase family 1 (GH1) beta-glucosidases play roles in many processes in plants, such as chemical defense, alkaloid metabolism, hydrolysis of cell wall-derived oligosaccharides, phytohormone regulation, and lignification. However, the functions of most of the 34 GH1 gene products in rice (Oryza sativa) are unknown. Os3BGlu6, a rice beta-glucosidase representing a previously uncharacterized phylogenetic cluster of GH1, was produced in recombinant Escherichia coli. Os3BGlu6 hydrolyzed p-nitrophenyl (pNP)-beta-d-fucoside (k(cat)/K(m) = 67 mm(-1) s(-1)), pNP-beta-d-glucoside (k(cat)/K(m) = 6.2 mm(-1) s(-1)), and pNP-beta-d-galactoside (k(cat)/K(m) = 1.6 mm(-1)s(-1)) efficiently but had little activity toward other pNP glycosides. It also had high activity toward n-octyl-beta-d-glucoside and beta-(1-->3)- and beta-(1-->2)-linked disaccharides and was able to hydrolyze apigenin beta-glucoside and several other natural glycosides. Crystal structures of Os3BGlu6 and its complexes with a covalent intermediate, 2-deoxy-2-fluoroglucoside, and a nonhydrolyzable substrate analog, n-octyl-beta-d-thioglucopyranoside, were solved at 1.83, 1.81, and 1.80 A resolution, respectively. The position of the covalently trapped 2-F-glucosyl residue in the enzyme was similar to that in a 2-F-glucosyl intermediate complex of Os3BGlu7 (rice BGlu1). The side chain of methionine-251 in the mouth of the active site appeared to block the binding of extended beta-(1-->4)-linked oligosaccharides and interact with the hydrophobic aglycone of n-octyl-beta-d-thioglucopyranoside. This correlates with the preference of Os3BGlu6 for short oligosaccharides and hydrophobic glycosides.
Figures

Similar articles
-
The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing β-glucosidase with significant thioglucohydrolase activity.Arch Biochem Biophys. 2011 Jun 1;510(1):62-72. doi: 10.1016/j.abb.2011.04.005. Epub 2011 Apr 16. Arch Biochem Biophys. 2011. PMID: 21521631
-
Enzymatic and structural characterization of hydrolysis of gibberellin A4 glucosyl ester by a rice β-D-glucosidase.Arch Biochem Biophys. 2013 Sep 1;537(1):39-48. doi: 10.1016/j.abb.2013.06.005. Epub 2013 Jun 26. Arch Biochem Biophys. 2013. PMID: 23811195
-
Structural insights into rice BGlu1 beta-glucosidase oligosaccharide hydrolysis and transglycosylation.J Mol Biol. 2008 Apr 4;377(4):1200-15. doi: 10.1016/j.jmb.2008.01.076. Epub 2008 Feb 4. J Mol Biol. 2008. PMID: 18308333
-
β-Glucosidases: Multitasking, moonlighting or simply misunderstood?Plant Sci. 2015 Dec;241:246-59. doi: 10.1016/j.plantsci.2015.10.014. Epub 2015 Oct 28. Plant Sci. 2015. PMID: 26706075 Review.
-
Dissecting the catalytic mechanism of a plant beta-D-glucan glucohydrolase through structural biology using inhibitors and substrate analogues.Carbohydr Res. 2007 Sep 3;342(12-13):1613-23. doi: 10.1016/j.carres.2007.05.013. Epub 2007 May 18. Carbohydr Res. 2007. PMID: 17548065 Review.
Cited by
-
Physiological and Transcriptome Analysis Provide Insights into the Effects of Low and High Selenium on Methionine and Starch Metabolism in Rice Seedlings.Int J Mol Sci. 2025 Feb 13;26(4):1596. doi: 10.3390/ijms26041596. Int J Mol Sci. 2025. PMID: 40004061 Free PMC article.
-
In Silico Analysis of the Molecular Interaction between Anthocyanase, Peroxidase and Polyphenol Oxidase with Anthocyanins Found in Cranberries.Int J Mol Sci. 2024 Sep 27;25(19):10437. doi: 10.3390/ijms251910437. Int J Mol Sci. 2024. PMID: 39408771 Free PMC article.
-
Purification, Characterization, and cDNA Cloning of a Prominent β-Glucosidase from the Gut of the Xylophagous Cockroach Panesthia angustipennis spadica.J Appl Glycosci (1999). 2016 Aug 20;63(3):51-59. doi: 10.5458/jag.jag.JAG-2016_006. eCollection 2016. J Appl Glycosci (1999). 2016. PMID: 34354483 Free PMC article.
-
Functional Characterization of CsBGlu12, a β-Glucosidase from Crocus sativus, Provides Insights into Its Role in Abiotic Stress through Accumulation of Antioxidant Flavonols.J Biol Chem. 2017 Mar 17;292(11):4700-4713. doi: 10.1074/jbc.M116.762161. Epub 2017 Jan 31. J Biol Chem. 2017. PMID: 28154174 Free PMC article.
-
The C-Domain of Oleuropein β-Glucosidase Assists in Protein Folding and Sequesters the Enzyme in Nucleus.Plant Physiol. 2017 Jul;174(3):1371-1383. doi: 10.1104/pp.17.00512. Epub 2017 May 8. Plant Physiol. 2017. PMID: 28483880 Free PMC article.
References
-
- Babcock GD, Esen A (1994) Substrate specificity of maize β-glucosidase. Plant Sci 101 31–39
-
- Bachem CW, van der Höven RS, De Bruijin SM, Vreugdenhil D, Zabeau M, Visser RG (1996) Visualization of differential gene expression using novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J 9 745–753 - PubMed
-
- Barrett T, Suresh CG, Tolley SP, Dodson EJ, Hughes MA (1995) The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure 3 951–960 - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340 783–795 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

