Central role of the oxygen-dependent degradation domain of Drosophila HIFalpha/Sima in oxygen-dependent nuclear export
- PMID: 19587118
- PMCID: PMC2735486
- DOI: 10.1091/mbc.e09-01-0038
Central role of the oxygen-dependent degradation domain of Drosophila HIFalpha/Sima in oxygen-dependent nuclear export
Abstract
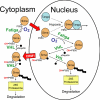
The Drosophila HIFalpha homologue, Sima, is localized mainly in the cytoplasm in normoxia and accumulates in the nucleus upon hypoxic exposure. We have characterized the mechanism governing Sima oxygen-dependent subcellular localization and found that Sima shuttles continuously between the nucleus and the cytoplasm. We have previously shown that nuclear import depends on an atypical bipartite nuclear localization signal mapping next to the C-terminus of the protein. We show here that nuclear export is mediated in part by a CRM1-dependent nuclear export signal localized in the oxygen-dependent degradation domain (ODDD). CRM1-dependent nuclear export requires both oxygen-dependent hydroxylation of a specific prolyl residue (Pro850) in the ODDD, and the activity of the von Hippel Lindau tumor suppressor factor. At high oxygen tension rapid nuclear export of Sima occurs, whereas in hypoxia, Sima nuclear export is largely inhibited. HIFalpha/Sima nucleo-cytoplasmic localization is the result of a dynamic equilibrium between nuclear import and nuclear export, and nuclear export is modulated by oxygen tension.
Figures





Similar articles
-
Regulation of the Drosophila hypoxia-inducible factor alpha Sima by CRM1-dependent nuclear export.Mol Cell Biol. 2008 May;28(10):3410-23. doi: 10.1128/MCB.01027-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332128 Free PMC article.
-
Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein.Mol Cell Biol. 2002 Aug;22(15):5319-36. doi: 10.1128/MCB.22.15.5319-5336.2002. Mol Cell Biol. 2002. PMID: 12101228 Free PMC article.
-
The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima.J Cell Sci. 2005 Dec 1;118(Pt 23):5431-41. doi: 10.1242/jcs.02648. Epub 2005 Nov 8. J Cell Sci. 2005. PMID: 16278294
-
Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing.J Mol Med (Berl). 2015 Jun;93(6):599-608. doi: 10.1007/s00109-015-1276-0. Epub 2015 Mar 27. J Mol Med (Berl). 2015. PMID: 25809665 Review.
-
Cellular and developmental adaptations to hypoxia: a Drosophila perspective.Methods Enzymol. 2007;435:123-44. doi: 10.1016/S0076-6879(07)35007-6. Methods Enzymol. 2007. PMID: 17998052 Review.
Cited by
-
Musashi mediates translational repression of the Drosophila hypoxia inducible factor.Nucleic Acids Res. 2016 Sep 19;44(16):7555-67. doi: 10.1093/nar/gkw372. Epub 2016 May 3. Nucleic Acids Res. 2016. PMID: 27141964 Free PMC article.
-
CRM1 blockade by selective inhibitors of nuclear export attenuates kidney cancer growth.J Urol. 2013 Jun;189(6):2317-26. doi: 10.1016/j.juro.2012.10.018. Epub 2012 Oct 16. J Urol. 2013. PMID: 23079374 Free PMC article.
-
Computational and experimental characterization of dVHL establish a Drosophila model of VHL syndrome.PLoS One. 2014 Oct 13;9(10):e109864. doi: 10.1371/journal.pone.0109864. eCollection 2014. PLoS One. 2014. PMID: 25310726 Free PMC article.
-
Longevity GWAS Using the Drosophila Genetic Reference Panel.J Gerontol A Biol Sci Med Sci. 2015 Dec;70(12):1470-8. doi: 10.1093/gerona/glv047. Epub 2015 Apr 28. J Gerontol A Biol Sci Med Sci. 2015. PMID: 25922346 Free PMC article.
-
A genetically encoded biosensor for visualising hypoxia responses in vivo.Biol Open. 2017 Feb 15;6(2):296-304. doi: 10.1242/bio.018226. Biol Open. 2017. PMID: 28011628 Free PMC article.
References
-
- Adryan B., Decker H. J., Papas T. S., Hsu T. Tracheal development and the von Hippel-Lindau tumor suppressor homolog in Drosophila. Oncogene. 2000;19:2803–2811. - PubMed
-
- Bacon N. C., Wappner P., O'Rourke J. F., Bartlett S. M., Shilo B., Pugh C. W., Ratcliffe P. J. Regulation of the Drosophila bHLH-PAS protein Sima by hypoxia: functional evidence for homology with mammalian HIF-1 alpha. Biochem. Biophys. Res. Commun. 1998;249:811–816. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

