The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice
- PMID: 19587447
- PMCID: PMC2719945
- DOI: 10.1172/JCI36908
The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice
Abstract
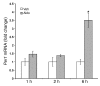
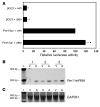
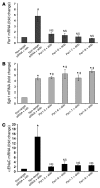
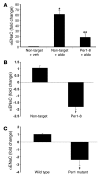
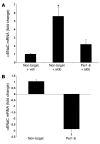
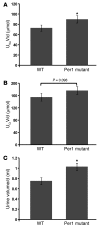
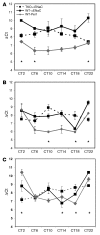
The mineralocorticoid aldosterone is a major regulator of sodium transport in target epithelia and contributes to the control of blood pressure and cardiac function. It specifically functions to increase renal absorption of sodium from tubular fluid via regulation of the alpha subunit of the epithelial sodium channel (alphaENaC). We previously used microarray technology to identify the immediate transcriptional targets of aldosterone in a mouse inner medullary collecting duct cell line and found that the transcript induced to the greatest extent was the circadian clock gene Period 1. Here, we investigated the role of Period 1 in mediating the downstream effects of aldosterone in renal cells. Aldosterone treatment stimulated expression of Period 1 (Per1) mRNA in renal collecting duct cell lines and in the rodent kidney. RNA silencing of Period 1 dramatically decreased expression of mRNA encoding alphaENaC in the presence or absence of aldosterone. Furthermore, expression of alphaENaC-encoding mRNA was attenuated in the renal medulla of mice with disruption of the Per1 gene, and these mice exhibited increased urinary sodium excretion. Renal alphaENaC-encoding mRNA was expressed in an apparent circadian pattern, and this pattern was dramatically altered in mice lacking functional Period genes. These results suggest a role for Period 1 in the regulation of the renal epithelial sodium channel and more broadly implicate the circadian clock in control of sodium balance.
Figures






References
-
- Booth R.E., Johnson J.P., Stockand J.D. Aldosterone. Adv. Physiol. Educ. 2002;26:8–20. - PubMed
-
- Gumz M.L., Popp M.P., Wingo C.S., Cain B.D. Early transcriptional effects of aldosterone in a mouse inner medullary collecting duct cell line. Am. J. Physiol. Renal Physiol. 2003;285:F664–F673. - PubMed
-
- Escoubet B., Coureau C., Bonvalet J.P., Farman N. Noncoordinate regulation of epithelial Na channel and Na pump subunit mRNAs in kidney and colon by aldosterone. Am. J. Physiol. 1997;272:C1482–C1491. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

