Chd1 regulates open chromatin and pluripotency of embryonic stem cells
- PMID: 19587682
- PMCID: PMC3891576
- DOI: 10.1038/nature08212
Chd1 regulates open chromatin and pluripotency of embryonic stem cells
Abstract
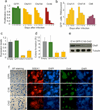
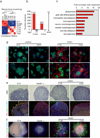
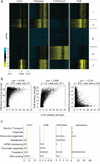
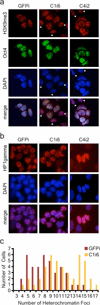
An open chromatin largely devoid of heterochromatin is a hallmark of stem cells. It remains unknown whether an open chromatin is necessary for the differentiation potential of stem cells, and which molecules are needed to maintain open chromatin. Here we show that the chromatin remodelling factor Chd1 is required to maintain the open chromatin of pluripotent mouse embryonic stem cells. Chd1 is a euchromatin protein that associates with the promoters of active genes, and downregulation of Chd1 leads to accumulation of heterochromatin. Chd1-deficient embryonic stem cells are no longer pluripotent, because they are incapable of giving rise to primitive endoderm and have a high propensity for neural differentiation. Furthermore, Chd1 is required for efficient reprogramming of fibroblasts to the pluripotent stem cell state. Our results indicate that Chd1 is essential for open chromatin and pluripotency of embryonic stem cells, and for somatic cell reprogramming to the pluripotent state.
Figures
Comment in
-
Stem cells: Escaping fates with open states.Nature. 2009 Aug 13;460(7257):802-3. doi: 10.1038/460802a. Nature. 2009. PMID: 19675633 No abstract available.
References
Bibliography
-
- Reddien P, Sanchez-Alvarado A. Fundamentals of Planarian Regenaration. Annu. Rev. Cell. Dev. Biol. 2004;20(1):725. - PubMed
-
- Terstappen LW, et al. Sequential generations of hematopoietic colonies derived from single nonlineage-committed CD34+CD38- progenitor cells. Blood. 1991;77(6):1218. - PubMed
-
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58. - PubMed
Methods Bibliography
-
- Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416(6880):545. - PubMed
-
- Reynolds A, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326. - PubMed
-
- Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

