A highly annotated whole-genome sequence of a Korean individual
- PMID: 19587683
- PMCID: PMC2860965
- DOI: 10.1038/nature08211
A highly annotated whole-genome sequence of a Korean individual
Abstract
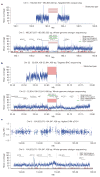
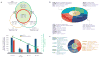
Recent advances in sequencing technologies have initiated an era of personal genome sequences. To date, human genome sequences have been reported for individuals with ancestry in three distinct geographical regions: a Yoruba African, two individuals of northwest European origin, and a person from China. Here we provide a highly annotated, whole-genome sequence for a Korean individual, known as AK1. The genome of AK1 was determined by an exacting, combined approach that included whole-genome shotgun sequencing (27.8x coverage), targeted bacterial artificial chromosome sequencing, and high-resolution comparative genomic hybridization using custom microarrays featuring more than 24 million probes. Alignment to the NCBI reference, a composite of several ethnic clades, disclosed nearly 3.45 million single nucleotide polymorphisms (SNPs), including 10,162 non-synonymous SNPs, and 170,202 deletion or insertion polymorphisms (indels). SNP and indel densities were strongly correlated genome-wide. Applying very conservative criteria yielded highly reliable copy number variants for clinical considerations. Potential medical phenotypes were annotated for non-synonymous SNPs, coding domain indels, and structural variants. The integration of several human whole-genome sequences derived from several ethnic groups will assist in understanding genetic ancestry, migration patterns and population bottlenecks.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

