Essential role for Schizosaccharomyces pombe pik1 in septation
- PMID: 19587793
- PMCID: PMC2704394
- DOI: 10.1371/journal.pone.0006179
Essential role for Schizosaccharomyces pombe pik1 in septation
Abstract
Background: Schizosaccharomyces pombe pik1 encodes a phosphatidylinositol 4-kinase, reported to bind Cdc4, but not Cdc4(G107S).
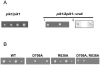
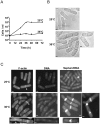
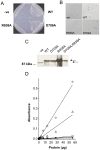
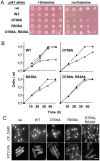
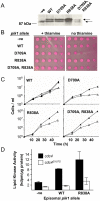
Principal findings: Gene deletion revealed that pik1 is essential. In cells with pik1 deleted, ectopic expression of a loss-of-function allele, created by fusion to a temperature-sensitive dihydrofolate reductase, allowed normal cell proliferation at 25 degrees C. At 36 degrees C, cells arrested with abnormally thick, misplaced or supernumerary septa, indicating a defect late in septation. In addition to being Golgi associated, ectopically expressed GFP-tagged Pik1 was observed at the medial cell plane late in cytokinesis. New alleles, created by site-directed mutagenesis, were expressed ectopically. Lipid kinase and Cdc4-binding activity assays were performed. Pik1(D709A) was kinase-dead, but bound Cdc4. Pik1(R838A) did not bind Cdc4, but was an active kinase. Genomic integration of these substitutions in S. pombe and complementation studies in Saccharomyces cerevisiae pik1-101 cells revealed that D709 is essential in both cases while R838 is dispensable. In S. pombe, ectopic expression of pik1 was dominantly lethal; while, pik1(D709A,R838A) was innocuous, pik1(R838A) was almost innocuous, and pik1(D709A) produced partial lethality and septation defects. The pik1 ectopic expression lethal phenotype was suppressed in cdc4(G107S). Thus, D709 is essential for kinase activity and septation.
Conclusions: Pik1 kinase activity is required for septation. The Pik1 R838 residue is required for important protein-protein interactions, possibly with Cdc4.
Conflict of interest statement
Figures





Similar articles
-
Cdc4p, a contractile ring protein essential for cytokinesis in Schizosaccharomyces pombe, interacts with a phosphatidylinositol 4-kinase.J Biol Chem. 2001 Feb 23;276(8):5932-42. doi: 10.1074/jbc.M008715200. Epub 2000 Nov 21. J Biol Chem. 2001. PMID: 11087749
-
Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis.J Cell Biol. 1995 Aug;130(3):651-60. doi: 10.1083/jcb.130.3.651. J Cell Biol. 1995. PMID: 7622565 Free PMC article.
-
Isolation of an HSP12-homologous gene of Schizosaccharomyces pombe suppressing a temperature-sensitive mutant allele of cdc4.Gene. 1996 Jun 12;172(1):125-9. doi: 10.1016/0378-1119(96)00058-3. Gene. 1996. PMID: 8654972
-
Mutation in fission yeast phosphatidylinositol 4-kinase Pik1 is synthetically lethal with defect in telomere protection protein Pot1.Biochem Biophys Res Commun. 2018 Feb 19;496(4):1284-1290. doi: 10.1016/j.bbrc.2018.02.001. Epub 2018 Feb 2. Biochem Biophys Res Commun. 2018. PMID: 29410177
-
Regulation of contractile ring formation and septation in Schizosaccharomyces pombe.Curr Opin Microbiol. 2015 Dec;28:46-52. doi: 10.1016/j.mib.2015.08.001. Epub 2015 Sep 3. Curr Opin Microbiol. 2015. PMID: 26340438 Free PMC article. Review.
Cited by
-
Pck2 association with the plasma membrane and efficient response of the cell integrity pathway require regulation of PI4P homeostasis by exomer.Open Biol. 2024 Nov;14(11):240101. doi: 10.1098/rsob.240101. Epub 2024 Nov 13. Open Biol. 2024. PMID: 39540318 Free PMC article.
-
Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans.Nat Commun. 2016 Sep 28;7:12766. doi: 10.1038/ncomms12766. Nat Commun. 2016. PMID: 27677328 Free PMC article.
-
Structure of a Ca2+-myristoyl switch protein that controls activation of a phosphatidylinositol 4-kinase in fission yeast.J Biol Chem. 2011 Apr 8;286(14):12565-77. doi: 10.1074/jbc.M110.208868. Epub 2011 Feb 2. J Biol Chem. 2011. PMID: 21288895 Free PMC article.
-
Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex.Cell Rep. 2014 Sep 11;8(5):1533-44. doi: 10.1016/j.celrep.2014.07.048. Epub 2014 Aug 21. Cell Rep. 2014. PMID: 25159149 Free PMC article.
-
Exploring Genetic Interactions with Telomere Protection Gene pot1 in Fission Yeast.Biomolecules. 2023 Feb 15;13(2):370. doi: 10.3390/biom13020370. Biomolecules. 2023. PMID: 36830739 Free PMC article. Review.
References
-
- De Matteis MA, Godi A. PI-loting membrane traffic. Nature cell biology. 2004;6(6):487–492. - PubMed
-
- Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Developmental cell. 2005;8(4):467–477. - PubMed
-
- Zhang Y, Sugiura R, Lu Y, Asami M, Maeda T, et al. Phosphatidylinositol 4-phosphate 5-kinase Its3 and calcineurin Ppb1 coordinately regulate cytokinesis in fission yeast. The Journal of biological chemistry. 2000;275(45):35600–35606. - PubMed
-
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends in cell biology. 2006;16(7):351–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

