Revealing system-level correlations between aging and calorie restriction using a mouse transcriptome
- PMID: 19590981
- PMCID: PMC2829640
- DOI: 10.1007/s11357-009-9106-3
Revealing system-level correlations between aging and calorie restriction using a mouse transcriptome
Abstract
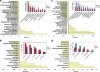
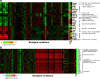
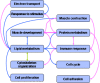
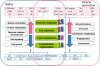
Although systems biology is a perfect framework for investigating system-level declines during aging, only a few reports have focused on a comprehensive understanding of system-level changes in the context of aging systems. The present study aimed to understand the most sensitive biological systems affected during aging and to reveal the systems underlying the crosstalk between aging and the ability of calorie restriction (CR) to effectively slow-down aging. We collected and analyzed 478 aging- and 586 CR-related mouse genes. For the given genes, the biological systems that are significantly related to aging and CR were examined according to three aspects. First, a global characterization by Gene Ontology (GO) was performed, where we found that the transcriptome (a set of genes) for both aging and CR were strongly related in the immune response, lipid metabolism, and cell adhesion functions. Second, the transcriptional modularity found in aging and CR was evaluated by identifying possible functional modules, sets of genes that show consistent expression patterns. Our analyses using the given functional modules, revealed systemic interactions among various biological processes, as exemplified by the negative relation shown between lipid metabolism and the immune response at the system level. Third, transcriptional regulatory systems were predicted for both the aging and CR transcriptomes. Here, we suggest a systems biology framework to further understand the most important systems as they age.
Figures
References
-
- Blalock EM, Chen KC, Stromberg AJ, Norris CM, Kadish I, Kraner SD, Porter NM, Landfield PW. Harnessing the power of gene microarrays for the study of brain aging and Alzheimer′s disease: statistical reliability and functional correlation. Ageing Res Rev. 2005;4:481–512. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
