Exploring functional roles of multibinding protein interfaces
- PMID: 19591200
- PMCID: PMC2776955
- DOI: 10.1002/pro.181
Exploring functional roles of multibinding protein interfaces
Abstract
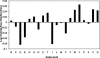
Cellular processes are highly interconnected and many proteins are shared in different pathways. Some of these shared proteins or protein families may interact with diverse partners using the same interface regions; such multibinding proteins are the subject of our study. The main goal of our study is to attempt to decipher the mechanisms of specific molecular recognition of multiple diverse partners by promiscuous protein regions. To address this, we attempt to analyze the physicochemical properties of multibinding interfaces and highlight the major mechanisms of functional switches realized through multibinding. We find that only 5% of protein families in the structure database have multibinding interfaces, and multibinding interfaces do not show any higher sequence conservation compared with the background interface sites. We highlight several important functional mechanisms utilized by multibinding families. (a) Overlap between different functional pathways can be prevented by the switches involving nearby residues of the same interfacial region. (b) Interfaces can be reused in pathways where the substrate should be passed from one protein to another sequentially. (c) The same protein family can develop different specificities toward different binding partners reusing the same interface; and finally, (d) inhibitors can attach to substrate binding sites as substrate mimicry and thereby prevent substrate binding.
Figures
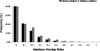
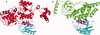
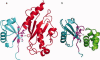
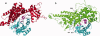
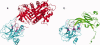
References
-
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
-
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt JF. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. - PubMed
-
- Krause R, von Mering C, Bork P, Dandekar T. Shared components of protein complexes—versatile building blocks or biochemical artefacts= Bioessays. 2004;26:1333–1343. - PubMed
-
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Beckett D. Functional switches in transcription regulation; molecular mimicry and plasticity in protein-protein interactions. Biochemistry. 2004;43:7983–7991. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

