Bioinformatic comparison of bacterial secretomes
- PMID: 19591790
- PMCID: PMC5054224
- DOI: 10.1016/S1672-0229(08)60031-5
Bioinformatic comparison of bacterial secretomes
Abstract
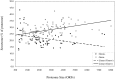
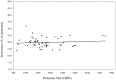
The rapid increasing number of completed bacterial genomes provides a good opportunity to compare their proteomes. This study was undertaken to specifically compare and contrast their secretomes-the fraction of the proteome with predicted N-terminal signal sequences, both type I and type II. A total of 176 theoretical bacterial proteomes were examined using the ExProt program. Compared with the Gram-positives, the Gram-negative bacteria were found, on average, to contain a larger number of potential Sec-dependent sequences. In the Gram-negative bacteria but not in the others, there was a positive correlation between proteome size and secretome size, while there was no correlation between secretome size and pathogenicity. Within the Gram-negative bacteria, intracellular pathogens were found to have the smallest secretomes. However, the secretomes of certain bacteria did not fit into the observed pattern. Specifically, the secretome of Borrelia burgdoferi has an unusually large number of putative lipoproteins, and the signal peptides of mycoplasmas show closer sequence similarity to those of the Gram-negative bacteria. Our analysis also suggests that even for a theoretical minimal genome of 300 open reading frames, a fraction of this gene pool (up to a maximum of 20%) may code for proteins with Sec-dependent signal sequences.
Figures

References
-
- Papanikou E. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 2007;5:839–851. - PubMed
-
- Saier M.H., Jr. Protein secretion and membrane insertion systems in Gram-negative bacteria. J. Membr. Biol. 2006;214:75–90. - PubMed
-
- Dalbey R.E., Kuhn A. Evolutionarily related insertion pathways of bacterial, mitochondrial, and thylakoid membrane proteins. Annu. Rev. Cell Dev. Biol. 2000;16:51–87. - PubMed
-
- Müller M., Klösgen R.B. The Tat pathway in bacteria and chloroplasts. Mol. Membr. Biol. 2005;22:113–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

