High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population
- PMID: 19591821
- PMCID: PMC2748313
- DOI: 10.1016/j.ydbio.2009.06.043
High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population
Abstract
The functional unit of the kidney is the nephron. During its organogenesis, the mammalian metanephric kidney generates thousands of nephrons over a protracted period of fetal life. All nephrons are derived from a population of self-renewing multi-potent progenitor cells, termed the cap mesenchyme. However, our understanding of the molecular and cellular mechanisms underlying nephron development is at an early stage. In order to identify factors involved in nephrogenesis, we performed a high-resolution, spatial profiling of a number of transcriptional regulators expressed within the cap mesenchyme and early developing nephron. Our results demonstrate novel, stereotypic, spatially defined cellular sub-domains within the cap mesenchyme, which may, in part, reflect induction of nephron precursors. These results suggest a hitherto unappreciated complexity of cell states that accompany the assembly of the metanephric kidney, likely reflecting diverse regulatory actions such as the maintenance and induction of nephron progenitors.
Figures



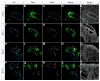
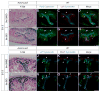

References
-
- Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–45. - PMC - PubMed
-
- Boyle S, Shioda T, Perantoni AO, de Caestecker M. Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn. 2007;236:2321–30. - PubMed
-
- Bridgewater D, Cox B, Cain J, Lau A, Athaide V, Gill PS, Kuure S, Sainio K, Rosenblum ND. Canonical WNT/beta-catenin signaling is required for ureteric branching. Dev Biol 2008 - PubMed
-
- Brodbeck S, Englert C. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

