T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire
- PMID: 19592274
- PMCID: PMC2965025
- DOI: 10.1016/j.immuni.2009.05.010
T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire
Abstract
Mouse type I natural killer T cell receptors (iNKT TCRs) use a single V alpha 14-J alpha 18 sequence and V beta s that are almost always V beta 8.2, V beta 7, or V beta 2, although the basis of this differential usage is unclear. We showed that the V beta bias occurred as a consequence of the CDR2 beta loops determining the affinity of the iNKT TCR for CD1d-glycolipids, thus controlling positive selection. Within a conserved iNKT-TCR-CD1d docking framework, these inherent V beta-CD1d affinities are further modulated by the hypervariable CDR3 beta loop, thereby defining a functional interplay between the two iNKT TCR CDR beta loops. These V beta biases revealed a broadly hierarchical response in which V beta 8.2 > V beta 7 > V beta 2 in the recognition of diverse CD1d ligands. This restriction of the iNKT TCR repertoire during thymic selection paradoxically ensures that each peripheral iNKT cell recognizes a similar spectrum of antigens.
Conflict of interest statement
The authors declare no financial conflict of interest.
Figures

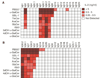
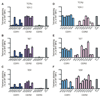
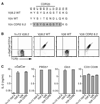

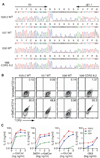
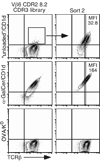
References
-
- Apostolou I, Cumano A, Gachelin G, Kourilsky P. Evidence for two subgroups of CD4−CD8− NKT cells with distinct TCRαβ repertoires and differential distribution in lymphoid tissues. J Immunol. 2000;165:2481–2490. - PubMed
-
- Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. - PubMed
-
- Behar SM, Podrebarac TA, Roy CJ, Wang CR, Brenner MB. Diverse TCRs recognize murine CD1. J Immunol. 1999;162:161–167. - PubMed
-
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 11331/CRUK_/Cancer Research UK/United Kingdom
- MC_U137884181/MRC_/Medical Research Council/United Kingdom
- R01 AI057485/AI/NIAID NIH HHS/United States
- AI18785/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- AI076463/AI/NIAID NIH HHS/United States
- R37 AI018785/AI/NIAID NIH HHS/United States
- AI078246/AI/NIAID NIH HHS/United States
- R21 AI076463/AI/NIAID NIH HHS/United States
- R56 AI078246/AI/NIAID NIH HHS/United States
- T32 AI07405/AI/NIAID NIH HHS/United States
- R56 AI018785/AI/NIAID NIH HHS/United States
- R01 AI057519/AI/NIAID NIH HHS/United States
- R01 AI018785/AI/NIAID NIH HHS/United States
- AI057485/AI/NIAID NIH HHS/United States
- P01 AI022295/AI/NIAID NIH HHS/United States
- AI057519/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

