An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4
- PMID: 19592493
- PMCID: PMC2757241
- DOI: 10.1074/jbc.M109.022392
An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4
Abstract
Toll-like receptors (TLRs) mediate responses to pathogen-associated molecules as part of the vertebrate innate immune response to infection. Receptor dimerization is coupled to downstream signal transduction by the recruitment of a post-receptor complex containing the adaptor protein MyD88 and the IRAK protein kinases. In this work, we show that the death domains of human MyD88 and IRAK-4 assemble into closed complexes having unusual stoichiometries of 7:4 and 8:4, the Myddosome. Formation of the Myddosome is likely to be a key event for TLR4 signaling in vivo as we show here that pathway activation requires that the receptors cluster into lipid rafts. Taken together, these findings indicate that TLR activation causes the formation of a highly oligomeric signaling platform analogous to the death-inducing signaling complex of the Fas receptor pathway.
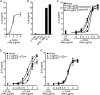
Figures




References
-
- Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 - PubMed
-
- Gay N. J., Gangloff M. (2007) Annu. Rev. Biochem. 76, 141–165 - PubMed
-
- O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 - PubMed
-
- Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) J. Biol. Chem. 273, 12203–12209 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

