Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects
- PMID: 19592581
- PMCID: PMC2742401
- DOI: 10.1093/hmg/ddp310
Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects
Abstract
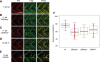
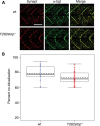
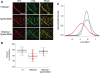
Spinal muscular atrophy (SMA), a recessive genetic disease, affects lower motoneurons leading to denervation, atrophy, paralysis and in severe cases death. Reduced levels of survival motor neuron (SMN) protein cause SMA. As a first step towards generating a genetic model of SMA in zebrafish, we identified three smn mutations. Two of these alleles, smnY262stop and smnL265stop, were stop mutations that resulted in exon 7 truncation, whereas the third, smnG264D, was a missense mutation corresponding to an amino acid altered in human SMA patients. Smn protein levels were low/undetectable in homozygous mutants consistent with unstable protein products. Homozygous mutants from all three alleles were smaller and survived on the basis of maternal Smn dying during the second week of larval development. Analysis of the neuromuscular system in these mutants revealed a decrease in the synaptic vesicle protein, SV2. However, two other synaptic vesicle proteins, synaptotagmin and synaptophysin were unaffected. To address whether the SV2 decrease was due specifically to Smn in motoneurons, we tested whether expressing human SMN protein exclusively in motoneurons in smn mutants could rescue the phenotype. For this, we generated a transgenic zebrafish line with human SMN driven by the motoneuron-specific zebrafish hb9 promoter and then generated smn mutant lines carrying this transgene. We found that introducing human SMN specifically into motoneurons rescued the SV2 decrease observed in smn mutants. Our analysis indicates the requirement for Smn in motoneurons to maintain SV2 in presynaptic terminals indicating that Smn, either directly or indirectly, plays a role in presynaptic integrity.
Figures




References
-
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Rochette C.F., Gilbert N., Simard L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. - PubMed
-
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. - PubMed
-
- Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

