Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD)
- PMID: 19593370
- PMCID: PMC2700282
- DOI: 10.1371/journal.pgen.1000559
Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD)
Abstract
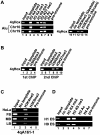
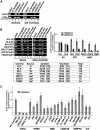
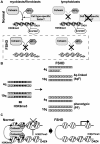
Facioscapulohumeral dystrophy (FSHD) is an autosomal dominant muscular dystrophy in which no mutation of pathogenic gene(s) has been identified. Instead, the disease is, in most cases, genetically linked to a contraction in the number of 3.3 kb D4Z4 repeats on chromosome 4q. How contraction of the 4qter D4Z4 repeats causes muscular dystrophy is not understood. In addition, a smaller group of FSHD cases are not associated with D4Z4 repeat contraction (termed "phenotypic" FSHD), and their etiology remains undefined. We carried out chromatin immunoprecipitation analysis using D4Z4-specific PCR primers to examine the D4Z4 chromatin structure in normal and patient cells as well as in small interfering RNA (siRNA)-treated cells. We found that SUV39H1-mediated H3K9 trimethylation at D4Z4 seen in normal cells is lost in FSHD. Furthermore, the loss of this histone modification occurs not only at the contracted 4q D4Z4 allele, but also at the genetically intact D4Z4 alleles on both chromosomes 4q and 10q, providing the first evidence that the genetic change (contraction) of one 4qD4Z4 allele spreads its effect to other genomic regions. Importantly, this epigenetic change was also observed in the phenotypic FSHD cases with no D4Z4 contraction, but not in other types of muscular dystrophies tested. We found that HP1gamma and cohesin are co-recruited to D4Z4 in an H3K9me3-dependent and cell type-specific manner, which is disrupted in FSHD. The results indicate that cohesin plays an active role in HP1 recruitment and is involved in cell type-specific D4Z4 chromatin regulation. Taken together, we identified the loss of both histone H3K9 trimethylation and HP1gamma/cohesin binding at D4Z4 to be a faithful marker for the FSHD phenotype. Based on these results, we propose a new model in which the epigenetic change initiated at 4q D4Z4 spreads its effect to other genomic regions, which compromises muscle-specific gene regulation leading to FSHD pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

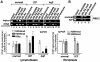



References
-
- Padberg GW. Facioscapulohumeral disease. [MD Thesis] Leiden, The Netherlands: Leiden University; 1982.
-
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. - PubMed
-
- Gabellini D, D'Antona G, Moggio M, Prelle A, Zecca C, et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. - PubMed
-
- Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

