A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana
- PMID: 19593375
- PMCID: PMC2700969
- DOI: 10.1371/journal.pgen.1000551
A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana
Abstract
Identifying natural allelic variation that underlies quantitative trait variation remains a fundamental problem in genetics. Most studies have employed either simple synthetic populations with restricted allelic variation or performed association mapping on a sample of naturally occurring haplotypes. Both of these approaches have some limitations, therefore alternative resources for the genetic dissection of complex traits continue to be sought. Here we describe one such alternative, the Multiparent Advanced Generation Inter-Cross (MAGIC). This approach is expected to improve the precision with which QTL can be mapped, improving the outlook for QTL cloning. Here, we present the first panel of MAGIC lines developed: a set of 527 recombinant inbred lines (RILs) descended from a heterogeneous stock of 19 intermated accessions of the plant Arabidopsis thaliana. These lines and the 19 founders were genotyped with 1,260 single nucleotide polymorphisms and phenotyped for development-related traits. Analytical methods were developed to fine-map quantitative trait loci (QTL) in the MAGIC lines by reconstructing the genome of each line as a mosaic of the founders. We show by simulation that QTL explaining 10% of the phenotypic variance will be detected in most situations with an average mapping error of about 300 kb, and that if the number of lines were doubled the mapping error would be under 200 kb. We also show how the power to detect a QTL and the mapping accuracy vary, depending on QTL location. We demonstrate the utility of this new mapping population by mapping several known QTL with high precision and by finding novel QTL for germination data and bolting time. Our results provide strong support for similar ongoing efforts to produce MAGIC lines in other organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
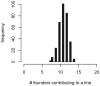
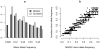
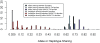
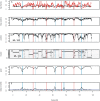
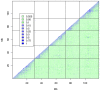
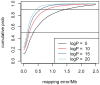
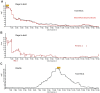
 for founder s at locus L is represented by a grey bar at coordinate (L,s), the shade of grey varying from white (P = 0) to black (P = 1). (E) The locus-specific power to detect a QTL explaining 10% of the phenotypic variance, from 40,000 simulations. In each simulation a 10% QTL was placed randomly along the genome. Successful detection is defined as the event that the genome-wide maximum in the genome scan for the QTL is within 3 Mb of the true QTL location. (F) The locus-specific median mapping error for the successfully detected QTL in (E).
for founder s at locus L is represented by a grey bar at coordinate (L,s), the shade of grey varying from white (P = 0) to black (P = 1). (E) The locus-specific power to detect a QTL explaining 10% of the phenotypic variance, from 40,000 simulations. In each simulation a 10% QTL was placed randomly along the genome. Successful detection is defined as the event that the genome-wide maximum in the genome scan for the QTL is within 3 Mb of the true QTL location. (F) The locus-specific median mapping error for the successfully detected QTL in (E).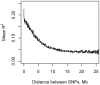
References
-
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int J Dev Biol. 2005;49:717–732. - PubMed
-
- Fernie AR, Tadmor Y, Zamir D. Natural genetic variation for improving crop quality. Curr Opin Plant Biol. 2006;9:196–202. - PubMed
-
- Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends in Plant Science. 2000;5:22–29. - PubMed
-
- Pigliucci M. Touchy and bushy: phenotypic plasticity and integration in response to wind stimulation in Arabidopsis thaliana. Int J Plant Sci. 2002;163:399–408.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

