Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking
- PMID: 19593404
- PMCID: PMC2707520
- DOI: 10.2119/molmed.2008.00130
Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking
Abstract
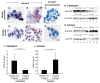
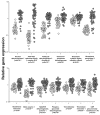
Nuclear factor erythroid 2-related factor 2 (Nrf2) is an oxidant-responsive transcription factor known to induce detoxifying and antioxidant genes. Cigarette smoke, with its large oxidant content, is a major stress on the cells of small airway epithelium, which are vulnerable to oxidant damage. We assessed the role of cigarette smoke in activation of Nrf2 in the human small airway epithelium in vivo. Fiberoptic bronchoscopy was used to sample the small airway epithelium in healthy-nonsmoker and healthy-smoker, and gene expression was assessed using microarrays. Relative to nonsmokers, Nrf2 protein in the small airway epithelium of smokers was activated and localized in the nucleus. The human homologs of 201 known murine Nrf2-modulated genes were identified, and 13 highly smoking-responsive Nrf2-modulated genes were identified. Construction of an Nrf2 index to assess the expression levels of these 13 genes in the airway epithelium of smokers showed coordinate control, an observation confirmed by quantitative PCR. This coordinate level of expression of the 13 Nrf2-modulated genes was independent of smoking history or demographic parameters. The Nrf2 index was used to identify two novel Nrf2-modulated, smoking-responsive genes, pirin (PIR) and UDP glucuronosyltransferase 1-family polypeptide A4 (UGT1A4). Both genes were demonstrated to contain functional antioxidant response elements in the promoter region. These observations suggest that Nrf2 plays an important role in regulating cellular defenses against smoking in the highly vulnerable small airway epithelium cells, and that there is variability within the human population in the Nrf2 responsiveness to oxidant burden.
Figures





Similar articles
-
Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis.Respir Res. 2007 Feb 8;8(1):10. doi: 10.1186/1465-9921-8-10. Respir Res. 2007. PMID: 17288615 Free PMC article.
-
Disparate oxidant gene expression of airway epithelium compared to alveolar macrophages in smokers.Respir Res. 2009 Nov 17;10(1):111. doi: 10.1186/1465-9921-10-111. Respir Res. 2009. PMID: 19919714 Free PMC article. Clinical Trial.
-
Cigarette smoking induces overexpression of a fat-depleting gene AZGP1 in the human.Chest. 2009 May;135(5):1197-1208. doi: 10.1378/chest.08-1024. Epub 2009 Feb 2. Chest. 2009. PMID: 19188554 Free PMC article.
-
Nrf2: friend and foe in preventing cigarette smoking-dependent lung disease.Chem Res Toxicol. 2012 Sep 17;25(9):1805-24. doi: 10.1021/tx300145n. Epub 2012 Jun 22. Chem Res Toxicol. 2012. PMID: 22686525 Review.
-
Nrf2 protects against airway disorders.Toxicol Appl Pharmacol. 2010 Apr 1;244(1):43-56. doi: 10.1016/j.taap.2009.07.024. Epub 2009 Jul 29. Toxicol Appl Pharmacol. 2010. PMID: 19646463 Review.
Cited by
-
EGF shifts human airway basal cell fate toward a smoking-associated airway epithelial phenotype.Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):12102-7. doi: 10.1073/pnas.1303058110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818594 Free PMC article.
-
Compartmentalization of anti-oxidant and anti-inflammatory gene expression in current and former smokers with COPD.Respir Res. 2019 Aug 20;20(1):190. doi: 10.1186/s12931-019-1164-1. Respir Res. 2019. PMID: 31429757 Free PMC article. Clinical Trial.
-
A comprehensive in silico exploration of the impacts of missense variants on two different conformations of human pirin protein.Bull Natl Res Cent. 2022;46(1):225. doi: 10.1186/s42269-022-00917-7. Epub 2022 Jul 30. Bull Natl Res Cent. 2022. PMID: 35967515 Free PMC article.
-
COPD lung studies of Nrf2 expression and the effects of Nrf2 activators.Inflammopharmacology. 2022 Aug;30(4):1431-1443. doi: 10.1007/s10787-022-00967-3. Epub 2022 Apr 20. Inflammopharmacology. 2022. PMID: 35441963 Free PMC article.
-
Interception of benzo[a]pyrene-7,8-dione by UDP glucuronosyltransferases (UGTs) in human lung cells.Chem Res Toxicol. 2013 Oct 21;26(10):1570-8. doi: 10.1021/tx400268q. Epub 2013 Oct 3. Chem Res Toxicol. 2013. PMID: 24047243 Free PMC article.
References
-
- Rahman I, MacNee W. Role of oxidants/antioxidants in smoking-induced lung diseases. Free Radic Biol Med. 1996;21:669–81. - PubMed
-
- MacNee W. Oxidants/antioxidants and COPD. Chest. 2000;117:303S–17S. - PubMed
-
- Rahman I, Biswas SK, Kode A. Oxidant and antioxidant balance in the airways and airway diseases. Eur J Pharmacol. 2006;533:222–39. - PubMed
-
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

