Sensitivity Measures for Oscillating Systems: Application to Mammalian Circadian Gene Network
- PMID: 19593456
- PMCID: PMC2707818
- DOI: 10.1109/TAC.2007.911364
Sensitivity Measures for Oscillating Systems: Application to Mammalian Circadian Gene Network
Abstract
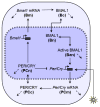
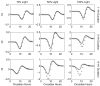
Vital physiological behaviors exhibited daily by bacteria, plants, and animals are governed by endogenous oscillators called circadian clocks. The most salient feature of the circadian clock is its ability to change its internal time (phase) to match that of the external environment. The circadian clock, like many oscillators in nature, is regulated at the cellular level by a complex network of interacting components. As a complementary approach to traditional biological investigation, we utilize mathematical models and systems theoretic tools to elucidate these mechanisms. The models are systems of ordinary differential equations exhibiting stable limit cycle behavior. To study the robustness of circadian phase behavior, we use sensitivity analysis. As the standard set of sensitivity tools are not suitable for the study of phase behavior, we introduce a novel tool, the parametric impulse phase response curve (pIPRC).
Figures





Similar articles
-
Mathematical Modeling in Circadian Rhythmicity.Methods Mol Biol. 2022;2482:55-80. doi: 10.1007/978-1-0716-2249-0_4. Methods Mol Biol. 2022. PMID: 35610419
-
Contribution of membrane-associated oscillators to biological timing at different timescales.Front Physiol. 2024 Jan 9;14:1243455. doi: 10.3389/fphys.2023.1243455. eCollection 2023. Front Physiol. 2024. PMID: 38264332 Free PMC article.
-
Regulation and function of extra-SCN circadian oscillators in the brain.Acta Physiol (Oxf). 2020 May;229(1):e13446. doi: 10.1111/apha.13446. Epub 2020 Feb 6. Acta Physiol (Oxf). 2020. PMID: 31965726 Review.
-
Entrainment of circadian clocks in mammals by arousal and food.Essays Biochem. 2011 Jun 30;49(1):119-36. doi: 10.1042/bse0490119. Essays Biochem. 2011. PMID: 21819388
-
Circadian clock in plants: Linking timing to fitness.J Integr Plant Biol. 2022 Apr;64(4):792-811. doi: 10.1111/jipb.13230. Epub 2022 Mar 5. J Integr Plant Biol. 2022. PMID: 35088570 Review.
Cited by
-
Weakly circadian cells improve resynchrony.PLoS Comput Biol. 2012;8(11):e1002787. doi: 10.1371/journal.pcbi.1002787. Epub 2012 Nov 29. PLoS Comput Biol. 2012. PMID: 23209395 Free PMC article.
-
Does the precision of a biological clock depend upon its period? Effects of the duper and tau mutations in Syrian hamsters.PLoS One. 2012;7(5):e36119. doi: 10.1371/journal.pone.0036119. Epub 2012 May 15. PLoS One. 2012. PMID: 22615753 Free PMC article.
-
A systems theoretic approach to analysis and control of mammalian circadian dynamics.Chem Eng Res Des. 2016 Dec;116:48-60. doi: 10.1016/j.cherd.2016.09.033. Epub 2016 Oct 8. Chem Eng Res Des. 2016. PMID: 28496287 Free PMC article.
-
Nonlinear Model Predictive Control For Circadian Entrainment Using Small-Molecule Pharmaceuticals.IFAC Pap OnLine. 2017 Jul;50(1):9864-9870. doi: 10.1016/j.ifacol.2017.08.1596. Epub 2017 Oct 18. IFAC Pap OnLine. 2017. PMID: 33842933 Free PMC article.
-
Systematic design methodology for robust genetic transistors based on I/O specifications via promoter-RBS libraries.BMC Syst Biol. 2013 Oct 27;7:109. doi: 10.1186/1752-0509-7-109. BMC Syst Biol. 2013. PMID: 24160305 Free PMC article.
References
-
- Sontag ED. Some new directions in control theory inspired by systems biology. IET Syst Biol. 2004 Jun;1(no. 1):9–18. [Online]. Available: http://dx.doi.org/10.1049/sb:20045006. - DOI - PubMed
-
- Kitano H. Systems biology: a brief overview. Proc Natl Acad Sci USA. 2002;295:1662–1664. [Online]. Available: http://dx.doi.org/10.1126/science.1069492. - DOI - PubMed
-
- Doyle FJ, III, Stelling J. Systems interface biology. J R Soc Interface. 2006 Oct;3(no. 10):603–616. [Online]. Available: http://dx.doi.org/10.1098/rsif.2006.0143. - DOI - PMC - PubMed
-
- Csete M, Doyle J. Reverse engineering of biological complexity. Proc Natl Acad Sci USA. 2002;295:1664–1669. [Online]. Available: http://dx.doi.org/10.1126/science.1069981. - DOI - PubMed
-
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, III, Doyle J. Robustness of cellular functions. Cell. 2004 Sep;118(no. 6):675–85. [Online]. Available: http://dx.doi.org/10.1016/j.cell.2004.09.008. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
