A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome
- PMID: 19594932
- PMCID: PMC2725114
- DOI: 10.1186/1471-2156-10-36
A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome
Abstract
Background: Most methods for constructing aneuploid yeast strains that have gained a specific chromosome rely on spontaneous failures of cell division fidelity. In Saccharomyces cerevisiae, extra chromosomes can be obtained when errors in meiosis or mitosis lead to nondisjunction, or when nuclear breakdown occurs in heterokaryons. We describe a strategy for constructing N+1 disomes that does not require such spontaneous failures. The method combines two well-characterized genetic tools: a conditional centromere that transiently blocks disjunction of one specific chromosome, and a duplication marker assay that identifies disomes among daughter cells. To test the strategy, we targeted chromosomes III, IV, and VI for duplication.
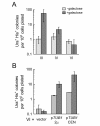
Results: The centromere of each chromosome was replaced by a centromere that can be blocked by growth in galactose, and ura3::HIS3, a duplication marker. Transient exposure to galactose induced the appearance of colonies carrying duplicated markers for chromosomes III or IV, but not VI. Microarray-based comparative genomic hybridization (CGH) confirmed that disomic strains carrying extra chromosome III or IV were generated. Chromosome VI contains several genes that are known to be deleterious when overexpressed, including the beta-tubulin gene TUB2. To test whether a tubulin stoichiometry imbalance is necessary for the apparent lethality caused by an extra chromosome VI, we supplied the parent strain with extra copies of the alpha-tubulin gene TUB1, then induced nondisjunction. Galactose-dependent chromosome VI disomes were produced, as revealed by CGH. Some chromosome VI disomes also carried extra, unselected copies of additional chromosomes.
Conclusion: This method causes efficient nondisjunction of a targeted chromosome and allows resulting disomic cells to be identified and maintained. We used the method to test the role of tubulin imbalance in the apparent lethality of disomic chromosome VI. Our results indicate that a tubulin imbalance is necessary for disomic VI lethality, but it may not be the only dosage-dependent effect.
Figures




References
-
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. - PubMed
-
- Cox BS, Bevan EA. Aneuploidy in yeast. New Phytologist. 1962;61:342–355.
-
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

