SR proteins in vertical integration of gene expression from transcription to RNA processing to translation
- PMID: 19595711
- PMCID: PMC2744344
- DOI: 10.1016/j.molcel.2009.06.016
SR proteins in vertical integration of gene expression from transcription to RNA processing to translation
Abstract
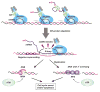
SR proteins have been studied extensively as a family of RNA-binding proteins that participate in both constitutive and regulated pre-mRNA splicing in mammalian cells. However, SR proteins were first discovered as factors that interact with transcriptionally active chromatin. Recent studies have now uncovered properties that connect these once apparently disparate functions, showing that a subset of SR proteins seem to bind directly to the histone 3 tail, play an active role in transcriptional elongation, and colocalize with genes that are engaged in specific intra- and interchromosome interactions for coordinated regulation of gene expression in the nucleus. These transcription-related activities are also coupled with a further expansion of putative functions of specific SR protein family members in RNA metabolism downstream of mRNA splicing, from RNA export to stability control to translation. These findings, therefore, highlight the broader roles of SR proteins in vertical integration of gene expression and provide mechanistic insights into their contributions to genome stability and proper cell-cycle progression in higher eukaryotic cells.
Figures

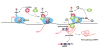

References
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Andersen FF, Tange TO, Sinnathamby T, Olesen JR, Andersen KE, Westergaard O, Kjems J, Knudsen BR. The RNA splicing factor ASF/SF2 inhibits human topoisomerase I mediated DNA relaxation. J Mol Biol. 2002;322:677–686. - PubMed
-
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Beyer AL, Osheim YN. Visualization of RNA transcription and processing. Semin Cell Biol. 1991;2:131–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

