Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases
- PMID: 19596236
- PMCID: PMC2861413
- DOI: 10.1016/j.cell.2009.06.029
Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases
Abstract
Structure-specific endonucleases resolve DNA secondary structures generated during DNA repair and recombination. The yeast 5' flap endonuclease Slx1-Slx4 has received particular attention with the finding that Slx4 has Slx1-independent key functions in genome maintenance. Although Slx1 is a highly conserved protein in eukaryotes, no orthologs of Slx4 were reported other than in fungi. Here we report the identification of Slx4 orthologs in metazoa, including fly MUS312, essential for meiotic recombination, and human BTBD12, an ATM/ATR checkpoint kinase substrate. Human SLX1-SLX4 displays robust Holliday junction resolvase activity in addition to 5' flap endonuclease activity. Depletion of SLX1 and SLX4 results in 53BP1 foci accumulation and H2AX phosphorylation as well as cellular hypersensitivity to MMS. Furthermore, we show that SLX4 binds the XPF(ERCC4) and MUS81 subunits of the XPF-ERCC1 and MUS81-EME1 endonucleases and is required for DNA interstrand crosslink repair. We propose that SLX4 acts as a docking platform for multiple structure-specific endonucleases.
Figures

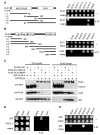
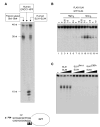




Comment in
-
Breaking up just got easier to do.Cell. 2009 Jul 10;138(1):20-2. doi: 10.1016/j.cell.2009.06.039. Cell. 2009. PMID: 19596231
References
-
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. - PubMed
-
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. - PubMed
-
- Bolt EL, Lloyd RG. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol Cell. 2002;10:187–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

