A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling
- PMID: 19596239
- PMCID: PMC2712736
- DOI: 10.1016/j.cell.2009.04.020
A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling
Abstract
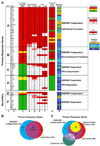
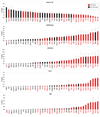
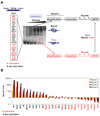
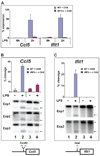
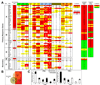
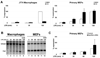
We describe a broad mechanistic framework for the transcriptional induction of mammalian primary response genes by Toll-like receptors and other stimuli. One major class of primary response genes is characterized by CpG-island promoters, which facilitate promiscuous induction from constitutively active chromatin without a requirement for SWI/SNF nucleosome remodeling complexes. The low nucleosome occupancy at promoters in this class can be attributed to the assembly of CpG islands into unstable nucleosomes, which may lead to SWI/SNF independence. Another major class consists of non-CpG-island promoters that assemble into stable nucleosomes, resulting in SWI/SNF dependence and a requirement for transcription factors that promote selective nucleosome remodeling. Some stimuli, including serum and tumor necrosis factor-alpha, exhibit a strong bias toward activation of SWI/SNF-independent CpG-island genes. In contrast, interferon-beta is strongly biased toward SWI/SNF-dependent non-CpG-island genes. By activating a diverse set of transcription factors, Toll-like receptors induce both classes and others for an optimal response to microbial pathogens.
Figures

Comment in
-
Teeing up transcription on CpG islands.Cell. 2009 Jul 10;138(1):14-6. doi: 10.1016/j.cell.2009.06.028. Cell. 2009. PMID: 19596228
-
The regulatory interplay of CpG islands and nucleosome remodeling at mammalian primary response genes.Epigenomics. 2009 Dec;1(2):233. Epigenomics. 2009. PMID: 22229158 No abstract available.
References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

