Control of inducible gene expression by signal-dependent transcriptional elongation
- PMID: 19596240
- PMCID: PMC2828818
- DOI: 10.1016/j.cell.2009.05.047
Control of inducible gene expression by signal-dependent transcriptional elongation
Abstract
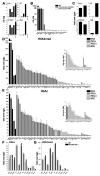
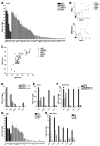
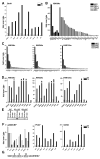
Most inducible transcriptional programs consist of primary and secondary response genes (PRGs and SRGs) that differ in their kinetics of expression and in their requirements for new protein synthesis and chromatin remodeling. Here we show that many PRGs, in contrast to SRGs, have preassembled RNA polymerase II (Pol II) and positive histone modifications at their promoters in the basal state. Pol II at PRGs generates low levels of full-length unspliced transcripts but fails to make mature, protein-coding transcripts in the absence of stimulation. Induction of PRGs is controlled at the level of transcriptional elongation and mRNA processing, through the signal-dependent recruitment of P-TEFb. P-TEFb is in turn recruited by the bromodomain-containing protein Brd4, which detects H4K5/8/12Ac inducibly acquired at PRG promoters. Our findings suggest that the permissive structure of PRGs both stipulates their unique regulation in the basal state by corepressor complexes and enables their rapid induction in multiple cell types.
Figures




Comment in
-
Teeing up transcription on CpG islands.Cell. 2009 Jul 10;138(1):14-6. doi: 10.1016/j.cell.2009.06.028. Cell. 2009. PMID: 19596228
References
-
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. - PubMed
-
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. - PubMed
-
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. - PubMed
-
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. - PubMed
-
- Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4:965–977. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

