CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence
- PMID: 19596241
- PMCID: PMC2756155
- DOI: 10.1016/j.cell.2009.04.041
CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence
Abstract
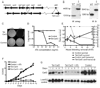
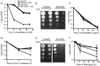
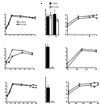
Mycobacterium tuberculosis is arguably the world's most successful infectious agent because of its ability to control its own cell growth within the host. Bacterial growth rate is closely coupled to rRNA transcription, which in E. coli is regulated through DksA and (p)ppGpp. The mechanisms of rRNA transcriptional control in mycobacteria, which lack DksA, are undefined. Here we identify CarD as an essential mycobacterial protein that controls rRNA transcription. Loss of CarD is lethal for mycobacteria in culture and during infection of mice. CarD depletion leads to sensitivity to killing by oxidative stress, starvation, and DNA damage, accompanied by failure to reduce rRNA transcription. CarD can functionally replace DksA for stringent control of rRNA transcription, even though CarD associates with a different site on RNA polymerase. These findings highlight a distinct molecular mechanism for regulating rRNA transcription in mycobacteria that is critical for M. tuberculosis pathogenesis.
Figures




References
-
- Alfoldi L, Stent GS, Clowes RC. The chromosomal site of the RNA control (RC) locus in Escherichia coli. J Mol Biol. 1962;5:348–355. - PubMed
-
- Avarbock D, Avarbock A, Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000;39:11640–11648. - PubMed
-
- Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. - PubMed
-
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. - PubMed
-
- Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., 3rd The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

