Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination
- PMID: 19596243
- PMCID: PMC2757090
- DOI: 10.1016/j.cell.2009.04.031
Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination
Abstract
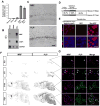
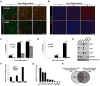
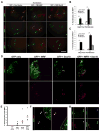
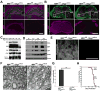
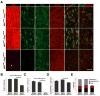
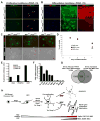
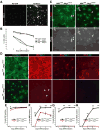
The transcriptional control of CNS myelin gene expression is poorly understood. Here we identify gene model 98, which we have named myelin gene regulatory factor (MRF), as a transcriptional regulator required for CNS myelination. Within the CNS, MRF is specifically expressed by postmitotic oligodendrocytes. MRF is a nuclear protein containing an evolutionarily conserved DNA binding domain homologous to a yeast transcription factor. Knockdown of MRF in oligodendrocytes by RNA interference prevents expression of most CNS myelin genes; conversely, overexpression of MRF within cultured oligodendrocyte progenitors or the chick spinal cord promotes expression of myelin genes. In mice lacking MRF within the oligodendrocyte lineage, premyelinating oligodendrocytes are generated but display severe deficits in myelin gene expression and fail to myelinate. These mice display severe neurological abnormalities and die because of seizures during the third postnatal week. These findings establish MRF as a critical transcriptional regulator essential for oligodendrocyte maturation and CNS myelination.
Figures
References
-
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. - PubMed
-
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, Raff MC. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. - PubMed
-
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. - PMC - PubMed
-
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

