Arginine methylation regulates telomere length and stability
- PMID: 19596784
- PMCID: PMC2738278
- DOI: 10.1128/MCB.00009-09
Arginine methylation regulates telomere length and stability
Abstract
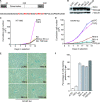
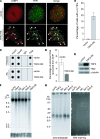
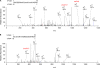
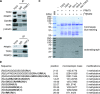
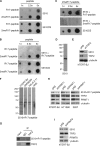
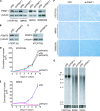
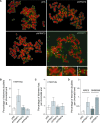
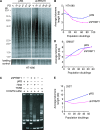
TRF2, a component of the shelterin complex, functions to protect telomeres. TRF2 contains an N-terminal basic domain rich in glycines and arginines, similar to the GAR motif that is methylated by protein arginine methyltransferases. However, whether arginine methylation regulates TRF2 function has not been determined. Here we report that amino acid substitutions of arginines with lysines in the basic domain of TRF2 induce telomere dysfunction-induced focus formation, leading to induction of cellular senescence. We have demonstrated that cells overexpressing TRF2 lysine mutants accumulate telomere doublets, indicative of telomere instability. We uncovered that TRF2 interacts with PRMT1, and its arginines in the basic domain undergo PRMT1-mediated methylation both in vitro and in vivo. We have shown that loss of PRMT1 induces growth arrest in normal human cells but has no effect on cell proliferation in cancer cells, suggesting that PRMT1 may control cell proliferation in a cell type-specific manner. We found that depletion of PRMT1 in normal human cells results in accumulation of telomere doublets, indistinguishable from overexpression of TRF2 lysine mutants. PRMT1 knockdown in cancer cells upregulates TRF2 association with telomeres, promoting telomere shortening. Taken together, these results suggest that PRMT1 may control telomere length and stability in part through TRF2 methylation.
Figures



References
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117735-748. - PubMed
-
- Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 223474-3487. - PMC - PubMed
-
- Bedford, M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 1204243-4246. - PubMed
-
- Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
