Mutation of asparagine 76 in the center of glutamine transporter SNAT3 modulates substrate-induced conductances and Na+ binding
- PMID: 19596860
- PMCID: PMC2757984
- DOI: 10.1074/jbc.M109.031013
Mutation of asparagine 76 in the center of glutamine transporter SNAT3 modulates substrate-induced conductances and Na+ binding
Abstract
The glutamine transporter SLC38A3 (SNAT3) plays an important role in the release of glutamine from brain astrocytes and the uptake of glutamine into hepatocytes. It is related to the vesicular GABA (gamma-aminobutyric acid) transporter and the SLC36 family of proton-amino acid cotransporters. The transporter carries out electroneutral Na+-glutamine cotransport-H+ antiport. In addition, substrate-induced uncoupled cation currents are observed. Mutation of asparagine 76 to glutamine or histidine in predicted transmembrane helix 1 abolished all substrate-induced currents. Mutation of asparagine 76 to aspartate rendered the transporter Na+-independent and resulted in a gain of a large substrate-induced chloride conductance in the absence of Na+. Thus, a single residue is critical for coupled and uncoupled ion flows in the glutamine transporter SNAT3. Homology modeling of SNAT3 along the structure of the related benzyl-hydantoin permease from Microbacterium liquefaciens reveals that Asn-76 is likely to be located in the center of the membrane close to the translocation pore and forms part of the predicted Na+ -binding site.
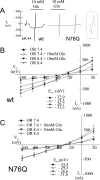
Figures







References
-
- Sundberg B. E., Wååg E., Jacobsson J. A., Stephansson O., Rumaks J., Svirskis S., Alsiö J., Roman E., Ebendal T., Klusa V., Fredriksson R. (2008) J. Mol. Neurosci. 35, 179–193 - PubMed
-
- Gasnier B. (2004) Pflugers Arch. 447, 756–759 - PubMed
-
- Boll M., Daniel H., Gasnier B. (2004) Pflugers Arch. 447, 776–779 - PubMed
-
- Mackenzie B., Erickson J. D. (2004) Pflugers Arch. 447, 784–795 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

