Temperature-sensitive mutations made easy: generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges
- PMID: 19596904
- PMCID: PMC2746138
- DOI: 10.1534/genetics.109.104794
Temperature-sensitive mutations made easy: generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges
Abstract
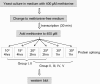
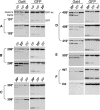
Reversible and easy to use, temperature-sensitive (TS) mutations are powerful tools for studying gene function. However, TS alleles are rare and difficult to generate and identify, and this has limited their use in most multicellular organisms. We have generated and characterized 41 intein switches, temperature-sensitive Sce VMA mutations that splice only at the permissive temperatures to generate intact host proteins. At nonpermissive temperatures, they fail to splice, resulting in a loss of function of the proteins in which they reside. By inserting an intein switch into a protein of interest, one can turn on and off the activities of the engineered protein with a simple temperature shift. The 41 TS inteins function in five different temperature ranges, with permissive temperatures ranging from 18 degrees to 30 degrees . This collection makes it possible to choose a TS-intein switch according to the optimal growth temperature of an organism or to suit a special experimental design.
Figures





References
-
- Adam, E., and F. B. Perler, 2002. Development of a positive genetic selection system for inhibition of protein splicing using mycobacterial inteins in Escherichia coli DNA gyrase subunit A. J. Mol. Microbiol. Biotechnol. 4: 479–487. - PubMed
-
- Anraku, Y., R. Mizutani and Y. Satow, 2005. Protein splicing: its discovery and structural insight into novel chemical mechanisms. IUBMB Life 57: 563–574. - PubMed
-
- Cadwell, R. C., and G. F. Joyce, 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2: 28–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

