Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae
- PMID: 19596907
- PMCID: PMC2746140
- DOI: 10.1534/genetics.109.103846
Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae
Abstract
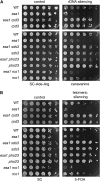
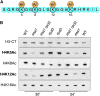
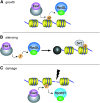
Histone modifications that regulate chromatin-dependent processes are catalyzed by multisubunit complexes. These can function in both targeting activities to specific genes and in regulating genomewide levels of modifications. In Saccharomyces cerevisiae, Esa1 and Rpd3 have opposing enzymatic activities and are catalytic subunits of multiple chromatin modifying complexes with key roles in processes such as transcriptional regulation and DNA repair. Esa1 is an essential histone acetyltransferase that belongs to the highly conserved MYST family. This study presents evidence that the yeast histone deacetylase gene, RPD3, when deleted, suppressed esa1 conditional mutant phenotypes. Deletion of RPD3 reversed rDNA and telomeric silencing defects and restored global H4 acetylation levels, in addition to rescuing the growth defect of a temperature-sensitive esa1 mutant. This functional genetic interaction between ESA1 and RPD3 was mediated through the Rpd3L complex. The suppression of esa1's growth defect by disruption of Rpd3L was dependent on lysine 12 of histone H4. We propose a model whereby Esa1 and Rpd3L act coordinately to control the acetylation of H4 lysine 12 to regulate transcription, thereby emphasizing the importance of dynamic acetylation and deacetylation of this particular histone residue in maintaining cell viability.
Figures




References
-
- Ahn, S. H., W. L. Cheung, J. Y. Hsu, R. L. Diaz, M. M. Smith et al., 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36. - PubMed
-
- Avvakumov, N., and J. Côté, 2007. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26: 5395–5407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

