Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection
- PMID: 19597497
- PMCID: PMC2908989
- DOI: 10.1038/ni.1769
Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection
Erratum in
- Nat Immunol. 2010 Jan;11(1):97
Abstract
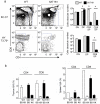
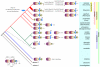
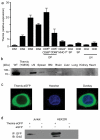
T cell antigen receptor (TCR) signaling in CD4(+)CD8(+) double-positive thymocytes determines cell survival and lineage commitment, but the genetic and molecular basis of this process is poorly defined. To address this issue, we used ethylnitrosourea mutagenesis to identify a previously unknown T lineage-specific gene, Themis, which is critical for the completion of positive selection. Themis contains a tandem repeat of a unique globular domain (called 'CABIT' here) that includes a cysteine motif that defines a family of five uncharacterized vertebrate proteins with orthologs in most animal species. Themis-deficient thymocytes showed no substantial impairment in early TCR signaling but did show altered expression of genes involved in the cell cycle and survival before and during positive selection. Our data suggest a unique function for Themis in sustaining positive selection.
Figures




Comment in
-
Themis imposes new law and order on positive selection.Nat Immunol. 2009 Aug;10(8):805-6. doi: 10.1038/ni0809-805. Nat Immunol. 2009. PMID: 19621038 No abstract available.
References
-
- He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. - PubMed
-
- Hughes P, Bouillet P, Strasser A. Role of Bim and other Bcl-2 family members in autoimmune and degenerative diseases. Curr Dir Autoimmun. 2006;9:74–94. - PubMed
-
- Mitchell BS, Kelley WN. Purinogenic immunodeficiency diseases: clinical features and molecular mechanisms. Ann Intern Med. 1980;92:826–831. - PubMed
-
- Taniuchi I, Littman DR. Epigenetic gene silencing by Runx proteins. Oncogene. 2004;23:4341–4345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

