A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints
- PMID: 19597943
- PMCID: PMC2824793
- DOI: 10.1007/s10858-009-9328-9
A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints
Abstract
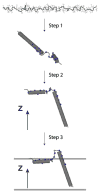
To fully describe the fold space and ultimately the biological function of membrane proteins, it is necessary to determine the specific interactions of the protein with the membrane. This property of membrane proteins that we refer to as structural topology cannot be resolved using X-ray crystallography or solution NMR alone. In this article, we incorporate into XPLOR-NIH a hybrid objective function for membrane protein structure determination that utilizes solution and solid-state NMR restraints, simultaneously defining structure, topology, and depth of insertion. Distance and angular restraints obtained from solution NMR of membrane proteins solubilized in detergent micelles are combined with backbone orientational restraints (chemical shift anisotropy and dipolar couplings) derived from solid-state NMR in aligned lipid bilayers. In addition, a supplementary knowledge-based potential, E (z) (insertion depth potential), is used to ensure the correct positioning of secondary structural elements with respect to a virtual membrane. The hybrid objective function is minimized using a simulated annealing protocol implemented into XPLOR-NIH software for general use.
Figures





Similar articles
-
Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach.Proc Natl Acad Sci U S A. 2009 Jun 23;106(25):10165-70. doi: 10.1073/pnas.0904290106. Epub 2009 Jun 9. Proc Natl Acad Sci U S A. 2009. PMID: 19509339 Free PMC article.
-
Solid-state NMR structures of integral membrane proteins.Mol Membr Biol. 2015 Aug-Dec;32(5-8):156-78. doi: 10.3109/09687688.2016.1139754. Epub 2016 Feb 8. Mol Membr Biol. 2015. PMID: 26857803 Review.
-
Multidimensional oriented solid-state NMR experiments enable the sequential assignment of uniformly 15N labeled integral membrane proteins in magnetically aligned lipid bilayers.J Biomol NMR. 2011 Nov;51(3):339-46. doi: 10.1007/s10858-011-9571-8. J Biomol NMR. 2011. PMID: 21976256
-
Solid-State NMR of Membrane Proteins in Lipid Bilayers: To Spin or Not To Spin?Acc Chem Res. 2021 Mar 16;54(6):1430-1439. doi: 10.1021/acs.accounts.0c00670. Epub 2021 Mar 3. Acc Chem Res. 2021. PMID: 33655754 Free PMC article. Review.
-
Atomic refinement with correlated solid-state NMR restraints.J Magn Reson. 2003 Aug;163(2):300-9. doi: 10.1016/s1090-7807(03)00147-2. J Magn Reson. 2003. PMID: 12914845
Cited by
-
Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach.Proc Natl Acad Sci U S A. 2009 Jun 23;106(25):10165-70. doi: 10.1073/pnas.0904290106. Epub 2009 Jun 9. Proc Natl Acad Sci U S A. 2009. PMID: 19509339 Free PMC article.
-
Probing membrane topology of the antimicrobial peptide distinctin by solid-state NMR spectroscopy in zwitterionic and charged lipid bilayers.Biochim Biophys Acta. 2011 Jan;1808(1):34-40. doi: 10.1016/j.bbamem.2010.08.008. Epub 2010 Aug 16. Biochim Biophys Acta. 2011. PMID: 20719234 Free PMC article.
-
A Practical Implicit Membrane Potential for NMR Structure Calculations of Membrane Proteins.Biophys J. 2015 Aug 4;109(3):574-85. doi: 10.1016/j.bpj.2015.06.047. Biophys J. 2015. PMID: 26244739 Free PMC article.
-
Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments.J Biol Chem. 2019 Nov 1;294(44):15914-15931. doi: 10.1074/jbc.REV119.009178. Epub 2019 Sep 24. J Biol Chem. 2019. PMID: 31551353 Free PMC article. Review.
-
Structural dynamics and conformational equilibria of SERCA regulatory proteins in membranes by solid-state NMR restrained simulations.Biophys J. 2014 Jun 17;106(12):2566-76. doi: 10.1016/j.bpj.2014.03.026. Biophys J. 2014. PMID: 24940774 Free PMC article.
References
-
- Bay DC, Rommens KL, Turner RJ. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta. 2008;1778:1814–1838. - PubMed
-
- Bertram R, Quine JR, Chapman MS, Cross TA. Atomic refinement using orientational restraints from solid-state NMR. J Magn Reson. 2000;147:9–16. - PubMed
-
- Buffy JJ, Traaseth NJ, Mascioni A, Gor’kov PL, Chekmenev EY, Brey WW, Veglia G. Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry. 2006;45:10939–10946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

