Instrumenting the health care enterprise for discovery research in the genomic era
- PMID: 19602638
- PMCID: PMC2752136
- DOI: 10.1101/gr.094615.109
Instrumenting the health care enterprise for discovery research in the genomic era
Abstract
Tens of thousands of subjects may be required to obtain reliable evidence relating disease characteristics to the weak effects typically reported from common genetic variants. The costs of assembling, phenotyping, and studying these large populations are substantial, recently estimated at three billion dollars for 500,000 individuals. They are also decade-long efforts. We hypothesized that automation and analytic tools can repurpose the informational byproducts of routine clinical care, bringing sample acquisition and phenotyping to the same high-throughput pace and commodity price-point as is currently true of genome-wide genotyping. Described here is a demonstration of the capability to acquire samples and data from densely phenotyped and genotyped individuals in the tens of thousands for common diseases (e.g., in a 1-yr period: N = 15,798 for rheumatoid arthritis; N = 42,238 for asthma; N = 34,535 for major depressive disorder) in one academic health center at an order of magnitude lower cost. Even for rare diseases caused by rare, highly penetrant mutations such as Huntington disease (N = 102) and autism (N = 756), these capabilities are also of interest.
Figures
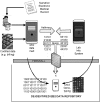

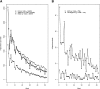


References
-
- Allison JJ, Wall TC, Spettell CM, Calhoun J, Fargason CA, Jr, Kobylinski RW, Farmer R, Kiefe C. The art and science of chart review. Jt Comm J Qual Improv. 2000;26:115–136. - PubMed
-
- Buyske S, Yang G, Matise T, Gordon D. When a case is not a case: Effects of phenotype misclassification on power and sample size requirements for the transmission disequilibrium test with affected child trios. Hum Hered. 2009;67:287–292. - PubMed
-
- Catlin A, Cowan C, Hartman M, Heffler S. National health spending in 2006: A year of change for prescription drugs. Health Aff. 2008;27:14–29. - PubMed
-
- Clayton PD, Boebert WE, Defriese GH, Dowell SP, Fennell ML, Frawley KA, Glaser J, Kemmerer RA, Landwehr CE, Rindfleisch TC, et al. For the Record: Protecting Electronic Health Information. National Academy Press; Washington, DC: 1997.
-
- Committee on Quality of Health Care in America, Institute of Medicine. Crossing the Quality Chasm: A New Health System for the 21st Century. National Academy Press; Washington, DC: 2001.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
