In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei
- PMID: 19602707
- PMCID: PMC2758762
- DOI: 10.1074/mcp.M900188-MCP200
In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei
Abstract
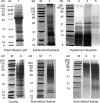
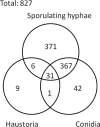
To further our understanding of powdery mildew biology during infection, we undertook a systematic shotgun proteomics analysis of the obligate biotroph Blumeria graminis f. sp. hordei at different stages of development in the host. Moreover we used a proteogenomics approach to feed information into the annotation of the newly sequenced genome. We analyzed and compared the proteomes from three stages of development representing different functions during the plant-dependent vegetative life cycle of this fungus. We identified 441 proteins in ungerminated spores, 775 proteins in epiphytic sporulating hyphae, and 47 proteins from haustoria inside barley leaf epidermal cells and used the data to aid annotation of the B. graminis f. sp. hordei genome. We also compared the differences in the protein complement of these key stages. Although confirming some of the previously reported findings and models derived from the analysis of transcriptome dynamics, our results also suggest that the intracellular haustoria are subject to stress possibly as a result of the plant defense strategy, including the production of reactive oxygen species. In addition, a number of small haustorial proteins with a predicted N-terminal signal peptide for secretion were identified in infected tissues: these represent candidate effector proteins that may play a role in controlling host metabolism and immunity.
Figures




References
-
- Marris E., Dodds P., Glover J., Hibberd J., Zhang J. H., Sayre R. (2008) Agronomy: five crop researchers who could change the world. Nature 456, 563–568 - PubMed
-
- Glawe D. A. (2008) The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol 46, 27–51 - PubMed
-
- Both M., Spanu P. (2004) Blumeria graminis f. sp. hordei, an obligate pathogen of barley, in Plant Pathogen Interactions (Talbot N. ed) pp. 202–218, Blackwell Publishing, Oxford
-
- Bushnell W. R. (1972) Physiology of fungal haustoria. Annu. Rev. Phytopathol 10, 151–176
-
- Catanzariti A. M., Dodds P. N., Ellis J. G. (2007) Avirulence proteins from haustoria-forming pathogens. FEMS Microbiol. Lett 269, 181–188 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

