Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis
- PMID: 19603075
- PMCID: PMC2705793
- DOI: 10.1371/journal.pone.0006072
Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis
Abstract
Background: Streptococcus suis is a zoonotic pathogen that infects pigs and can occasionally cause serious infections in humans. S. suis infections occur sporadically in human Europe and North America, but a recent major outbreak has been described in China with high levels of mortality. The mechanisms of S. suis pathogenesis in humans and pigs are poorly understood.
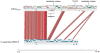
Methodology/principal findings: The sequencing of whole genomes of S. suis isolates provides opportunities to investigate the genetic basis of infection. Here we describe whole genome sequences of three S. suis strains from the same lineage: one from European pigs, and two from human cases from China and Vietnam. Comparative genomic analysis was used to investigate the variability of these strains. S. suis is phylogenetically distinct from other Streptococcus species for which genome sequences are currently available. Accordingly, approximately 40% of the approximately 2 Mb genome is unique in comparison to other Streptococcus species. Finer genomic comparisons within the species showed a high level of sequence conservation; virtually all of the genome is common to the S. suis strains. The only exceptions are three approximately 90 kb regions, present in the two isolates from humans, composed of integrative conjugative elements and transposons. Carried in these regions are coding sequences associated with drug resistance. In addition, small-scale sequence variation has generated pseudogenes in putative virulence and colonization factors.
Conclusions/significance: The genomic inventories of genetically related S. suis strains, isolated from distinct hosts and diseases, exhibit high levels of conservation. However, the genomes provide evidence that horizontal gene transfer has contributed to the evolution of drug resistance.
Conflict of interest statement
Figures



References
-
- Gottschalk M, Segura M, Xu J. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev. 2007;8:29–45. - PubMed
-
- Perch B, Kristjansen P, Skadhauge K. Group R streptococci pathogenic for man. Two cases of meningitis and one fatal case of sepsis. Acta Pathol Microbiol Scand. 1968;74:69–76. - PubMed
-
- Mai NT, Hoa NT, Nga TV, Linh LD, Chau TT, et al. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. 2008;46:659–667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

