Dynamin2 GTPase and cortactin remodel actin filaments
- PMID: 19605363
- PMCID: PMC2781994
- DOI: 10.1074/jbc.M109.024398
Dynamin2 GTPase and cortactin remodel actin filaments
Abstract
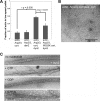
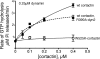
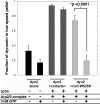
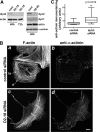
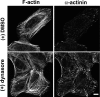
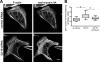
The large GTPase dynamin, best known for its activities that remodel membranes during endocytosis, also regulates F-actin-rich structures, including podosomes, phagocytic cups, actin comet tails, subcortical ruffles, and stress fibers. The mechanisms by which dynamin regulates actin filaments are not known, but an emerging view is that dynamin influences F-actin via its interactions with proteins that interact directly or indirectly with actin filaments. We show here that dynamin2 GTPase activity remodels actin filaments in vitro via a mechanism that depends on the binding partner and F-actin-binding protein, cortactin. Tightly associated actin filaments cross-linked by dynamin2 and cortactin became loosely associated after GTP addition when viewed by transmission electron microscopy. Actin filaments were dynamically unraveled and fragmented after GTP addition when viewed in real time using total internal reflection fluorescence microscopy. Cortactin stimulated the intrinsic GTPase activity of dynamin2 and maintained a stable link between actin filaments and dynamin2, even in the presence of GTP. Filaments remodeled by dynamin2 GTPase in vitro exhibit enhanced sensitivity to severing by the actin depolymerizing factor, cofilin, suggesting that GTPase-dependent remodeling influences the interactions of actin regulatory proteins and F-actin. The global organization of the actomyosin cytoskeleton was perturbed in U2-OS cells depleted of dynamin2, implicating dynamin2 in remodeling actin filaments that comprise supramolecular F-actin arrays in vivo. We conclude that dynamin2 GTPase remodels actin filaments and plays a role in orchestrating the global actomyosin cytoskeleton.
Figures


References
-
- Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 - PubMed
-
- Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. (1999) Nature 401, 613–616 - PubMed
-
- Upadhyaya A., van Oudenaarden A. (2003) Curr. Biol. 13, R734–R744 - PubMed
-
- Orth J. D., McNiven M. A. (2003) Curr. Opin. Cell Biol. 15, 31–39 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

