The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development
- PMID: 19607824
- PMCID: PMC2760734
- DOI: 10.1016/j.ydbio.2009.07.003
The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development
Abstract
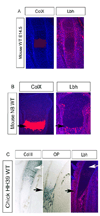
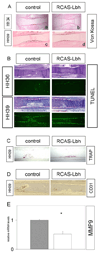
Lbh is thought to act as a transcriptional cofactor and is highly conserved among species. Here we show that Lbh is expressed in chondrocytes, cells of the perichondrium, and the primary spongiosa in fetal growth plates of mice and chickens. Lbh overexpression in chick wings, using the RCAS-retroviral vector strategy, results in shortened skeletal elements and delayed hypertrophic chondrocyte maturation and bone formation. Additionally, osteoclast and endothelial cell invasion are delayed in the Lbh-overexpressing bones. Finally, we find a dramatic suppression of Runx2 and VEGF mRNAs in chondrocytes and osteoblasts that overexpress Lbh. Strikingly, this abnormal bone development in infected limbs can be rescued by concurrent overexpression of Runx2. These results suggest that during endochondral bone formation, Lbh may negatively regulate vascular invasion and formation of the early ossification center at least in part by interfering with Runx2 and/or VEGF expression.
Figures




References
-
- Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:63–72. - PubMed
-
- Briegel KJ, Baldwin HS, Epstein JA, Joyner AL. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development. 2005;132:3305–3316. - PubMed
-
- Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233:291–304. - PubMed
-
- Carlevaro MF, Cermelli S, Cancedda R, Descalzi Cancedda F. Vascular endothelial growth factor (VEGF) in cartilage neovascularization and chondrocyte differentiation: auto-paracrine role during endochondral bone formation. J Cell Sci. 2000;113(Pt 1):59–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

