Prokaryotic ubiquitin-like protein pup is intrinsically disordered
- PMID: 19607839
- PMCID: PMC2734869
- DOI: 10.1016/j.jmb.2009.07.018
Prokaryotic ubiquitin-like protein pup is intrinsically disordered
Abstract
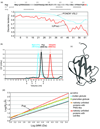
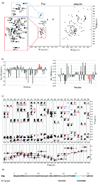
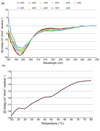
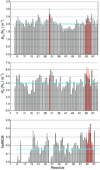
The prokaryotic ubiquitin-like protein Pup targets substrates for degradation by the Mycobacterium tuberculosis proteasome through its interaction with Mpa, an ATPase that is thought to abut the 20S catalytic subunit. Ubiquitin, which is assembled into a polymer to similarly signal for proteasomal degradation in eukaryotes, adopts a stable and compact structural fold that is adapted into other proteins for diverse biological functions. We used NMR spectroscopy to demonstrate that, unlike ubiquitin, the 64-amino-acid protein Pup is intrinsically disordered with small helical propensity in the C-terminal region. We found that the Pup:Mpa interaction involves an extensive contact surface that spans S21-K61 and that the binding is in the "slow exchange" regime on the NMR time scale, thus demonstrating higher affinity than most ubiquitin:ubiquitin receptor pairs. Interestingly, during the titration experiment, intermediate Pup species were observable, suggesting the formation of one or more transient state(s) upon binding. Moreover, Mpa selected one configuration for a region undergoing chemical exchange in the free protein. These findings provide mechanistic insights into Pup's functional role as a degradation signal.
Figures
References
-
- Zwickl P, Baumeister W, Steven A. Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–250. - PubMed
-
- Pouch MN, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol Microbiol. 2000;35:368–377. - PubMed
-
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. - PubMed
-
- Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

