Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry
- PMID: 19608599
- PMCID: PMC2773710
- DOI: 10.1074/mcp.M900317-MCP200
Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry
Abstract
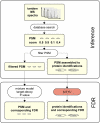
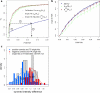
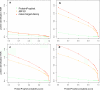
Comprehensive characterization of a proteome is a fundamental goal in proteomics. To achieve saturation coverage of a proteome or specific subproteome via tandem mass spectrometric identification of tryptic protein sample digests, proteomics data sets are growing dramatically in size and heterogeneity. The trend toward very large integrated data sets poses so far unsolved challenges to control the uncertainty of protein identifications going beyond well established confidence measures for peptide-spectrum matches. We present MAYU, a novel strategy that reliably estimates false discovery rates for protein identifications in large scale data sets. We validated and applied MAYU using various large proteomics data sets. The data show that the size of the data set has an important and previously underestimated impact on the reliability of protein identifications. We particularly found that protein false discovery rates are significantly elevated compared with those of peptide-spectrum matches. The function provided by MAYU is critical to control the quality of proteome data repositories and thereby to enhance any study relying on these data sources. The MAYU software is available as standalone software and also integrated into the Trans-Proteomic Pipeline.
Figures



References
-
- Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422,198–207 - PubMed
-
- Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Heck A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25,576–583 - PubMed
-
- Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125,187–199 - PubMed
-
- Omenn G. S., States D. J., Adamski M., Blackwell T. W., Menon R., Hermjakob H., Apweiler R., Haab B. B., Simpson R. J., Eddes J. S., Kapp E. A., Moritz R. L., Chan D. W., Rai A. J., Admon A., Aebersold R., Eng J., Hancock W. S., Hefta S. A., Meyer H., Paik Y. K., Yoo J. S., Ping P., Pounds J., Adkins J., Qian X., Wang R., Wasinger V., Wu C. Y., Zhao X., Zeng R., Archakov A., Tsugita A., Beer I., Pandey A., Pisano M., Andrews P., Tammen H., Speicher D. W., Hanash S. M. (2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 5,3226–3245 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

