A co-operative evaluation of different methods of detecting BCR-ABL kinase domain mutations in patients with chronic myeloid leukemia on second-line dasatinib or nilotinib therapy after failure of imatinib
- PMID: 19608684
- PMCID: PMC2738714
- DOI: 10.3324/haematol.2009.006981
A co-operative evaluation of different methods of detecting BCR-ABL kinase domain mutations in patients with chronic myeloid leukemia on second-line dasatinib or nilotinib therapy after failure of imatinib
Abstract
Background: Various techniques have been employed to detect BCR-ABL kinase domain mutations in patients with chronic myeloid leukemia who are resistant to imatinib. This has led to different reported frequencies of mutations and the finding of a heterogeneous pattern of individual mutations.
Design and methods: We compared direct sequencing alone and in combination with denaturing high-performance liquid chromatography and two high-sensitivity allele-specific oligonucleotide polymerase chain reaction approaches for analysis of BCR-ABL mutations in 200 blinded cDNA samples prior to and during second-line dasatinib or nilotinib therapy in patients with chronic myeloid leukemia in whom imatinib treatment had failed.
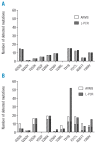
Results: One hundred and fourteen mutations were detected by both direct sequencing alone or in combination with high performance liquid chromatography and 13 mutations were additionally detected by the combined technique. Eighty of 83 mutations (96%) within a selected panel of 11 key mutations were confirmed by both allele-specific oligonucleotide polymerase chain reaction techniques and 62 mutations were identified in addition to those detected by combined liquid chromatography and direct sequencing, indicating the presence and a high prevalence of low-level mutations in this cohort of patients. Furthermore, 125 mutations were detected by only one allele-specific oligonucleotide polymerase chain reaction technique. Pre-existing mutations were traceable 4.5 months longer and emerging clones were detectable 3.0 months earlier by allele-specific oligonucleotide polymerase chain reaction than by direct sequencing together with liquid chromatography.
Conclusions: Our results suggest that denaturing high performance liquid chromatography combined with direct sequencing is a reliable screening technique for the detection of BCR-ABL kinase domain mutations. Allele-specific oligonucleotide polymerase chain reaction further increases the number of detected mutations and indicates a high prevalence of mutations at a low level. The clinical impact of such low-level mutations remains uncertain and requires further investigation. Allele-specific oligonucleotide polymerase chain reaction allows detection of defined mutations at a lower level than does denaturing high performance liquid chromatography combined with direct sequencing and may, therefore, provide clinical benefit by permitting early reconsideration of therapeutic strategies.
Figures


References
-
- Hehlmann R, Hochhaus A, Baccarani M European LeukemiaNet. Chronic myeloid leukaemia. Lancet. 2007;370:342–50. - PubMed
-
- O’Brien SG, Guilhot F, Goldman JM, Hochhaus A, Hughes TP, Radich JP, et al. International randomized study of interferon versus STI571 (IRIS) 7-year follow-up: sustained survival, low rate of transformation and increased rate of major molecular response (MMR) in patients (pts) with newly diagnosed chronic myeloid leukemia in chronic phase (CML-CP) treated with imatinib (IM) Blood. 2008;112 [abstract #186]
-
- Hochhaus A, Erben P, Ernst T, Mueller MC. Resistance to targeted therapy in chronic myelogenous leukemia. Semin Hematol. 2007;44 (Suppl 1):S15–24. - PubMed
-
- O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–9. - PubMed
-
- Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

